A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host
- PMID: 32632014
- PMCID: PMC7382252
- DOI: 10.1073/pnas.2003857117
A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host
Abstract
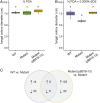
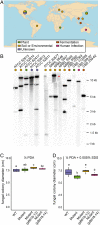
Symbioses of bacteria with fungi have only recently been described and are poorly understood. In the symbiosis of Mycetohabitans (formerly Burkholderia) rhizoxinica with the fungus Rhizopus microsporus, bacterial type III (T3) secretion is known to be essential. Proteins resembling T3-secreted transcription activator-like (TAL) effectors of plant pathogenic bacteria are encoded in the three sequenced Mycetohabitans spp. genomes. TAL effectors nuclear-localize in plants, where they bind and activate genes important in disease. The Burkholderia TAL-like (Btl) proteins bind DNA but lack the N- and C-terminal regions, in which TAL effectors harbor their T3 and nuclear localization signals, and activation domain. We characterized a Btl protein, Btl19-13, and found that, despite the structural differences, it can be T3-secreted and can nuclear-localize. A btl19-13 gene knockout did not prevent the bacterium from infecting the fungus, but the fungus became less tolerant to cell membrane stress. Btl19-13 did not alter transcription in a plant-based reporter assay, but 15 R. microsporus genes were differentially expressed in comparisons both of the fungus infected with the wild-type bacterium vs. the mutant and with the mutant vs. a complemented strain. Southern blotting revealed btl genes in 14 diverse Mycetohabitans isolates. However, banding patterns and available sequences suggest variation, and the btl19-13 phenotype could not be rescued by a btl gene from a different strain. Our findings support the conclusion that Btl proteins are effectors that act on host DNA and play important but varied or possibly host genotype-specific roles in the M. rhizoxinica-R. microsporus symbiosis.
Keywords: Btl proteins; Rhizopus microsporus; TAL effector; symbiosis; type III secretion.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

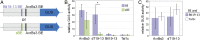
Similar articles
-
Prevalence and diversity of TAL effector-like proteins in fungal endosymbiotic Mycetohabitans spp.Microb Genom. 2024 Jun;10(6):001261. doi: 10.1099/mgen.0.001261. Microb Genom. 2024. PMID: 38860878 Free PMC article.
-
Evolution of an endofungal lifestyle: Deductions from the Burkholderia rhizoxinica genome.BMC Genomics. 2011 May 4;12:210. doi: 10.1186/1471-2164-12-210. BMC Genomics. 2011. PMID: 21539752 Free PMC article.
-
Transcription activator-like effectors from endosymbiotic bacteria control the reproduction of their fungal host.mBio. 2023 Dec 19;14(6):e0182423. doi: 10.1128/mbio.01824-23. Epub 2023 Nov 16. mBio. 2023. PMID: 37971247 Free PMC article.
-
Nodulation outer proteins: double-edged swords of symbiotic rhizobia.Biochem J. 2015 Sep 15;470(3):263-74. doi: 10.1042/BJ20150518. Biochem J. 2015. PMID: 26341483 Review.
-
TAL effectors are remote controls for gene activation.Curr Opin Microbiol. 2011 Feb;14(1):47-53. doi: 10.1016/j.mib.2010.12.001. Epub 2011 Jan 5. Curr Opin Microbiol. 2011. PMID: 21215685 Review.
Cited by
-
Let's Get Physical: Bacterial-Fungal Interactions and Their Consequences in Agriculture and Health.J Fungi (Basel). 2020 Oct 23;6(4):243. doi: 10.3390/jof6040243. J Fungi (Basel). 2020. PMID: 33114069 Free PMC article. Review.
-
A single-nucleotide insertion in Rxp confers durable resistance to bacterial pustule in soybean.Theor Appl Genet. 2024 Oct 23;137(11):254. doi: 10.1007/s00122-024-04743-5. Theor Appl Genet. 2024. PMID: 39441215
-
Molecular mechanisms that govern infection and antifungal resistance in Mucorales.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0018822. doi: 10.1128/mmbr.00188-22. Epub 2024 Mar 6. Microbiol Mol Biol Rev. 2024. PMID: 38445820 Free PMC article. Review.
-
Endofungal bacteria as hidden facilitators of biotic interactions.ISME J. 2025 Jan 2;19(1):wraf128. doi: 10.1093/ismejo/wraf128. ISME J. 2025. PMID: 40581745 Free PMC article. Review.
-
Symbioses between fungi and bacteria: from mechanisms to impacts on biodiversity.Curr Opin Microbiol. 2024 Aug;80:102496. doi: 10.1016/j.mib.2024.102496. Epub 2024 Jun 13. Curr Opin Microbiol. 2024. PMID: 38875733 Free PMC article. Review.
References
-
- Arora P., Riyaz-Ul-Hassan S., Endohyphal bacteria; The prokaryotic modulators of host fungal biology. Fungal Biol. Rev. 33, 72–81 (2018).
-
- Estrada-de Los Santos P. et al. ., Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel) 9, 389 (2018). - PMC - PubMed
-
- Partida-Martinez L. P., Hertweck C., Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 (2005). - PubMed
-
- Partida-Martinez L. P., Monajembashi S., Greulich K.-O., Hertweck C., Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17, 773–777 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

