A molecular mechanism for probabilistic bet hedging and its role in viral latency
- PMID: 32632017
- PMCID: PMC7382263
- DOI: 10.1073/pnas.1914430117
A molecular mechanism for probabilistic bet hedging and its role in viral latency
Abstract
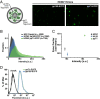
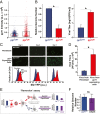
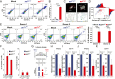
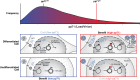
Probabilistic bet hedging, a strategy to maximize fitness in unpredictable environments by matching phenotypic variability to environmental variability, is theorized to account for the evolution of various fate-specification decisions, including viral latency. However, the molecular mechanisms underlying bet hedging remain unclear. Here, we report that large variability in protein abundance within individual herpesvirus virion particles enables probabilistic bet hedging between viral replication and latency. Superresolution imaging of individual virions of the human herpesvirus cytomegalovirus (CMV) showed that virion-to-virion levels of pp71 tegument protein-the major viral transactivator protein-exhibit extreme variability. This super-Poissonian tegument variability promoted alternate replicative strategies: high virion pp71 levels enhance viral replicative fitness but, strikingly, impede silencing, whereas low virion pp71 levels reduce fitness but promote silencing. Overall, the results indicate that stochastic tegument packaging provides a mechanism enabling probabilistic bet hedging between viral replication and latency.
Keywords: fate selection; herpesvirus; latency; stochastic variability; tegument.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Cohen D., Optimizing reproduction in a randomly varying environment. J. Theor. Biol. 12, 119–129 (1966). - PubMed
-
- Balaban N. Q., Persistence: Mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775 (2011). - PubMed
-
- Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S., Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

