Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia
- PMID: 32632085
- PMCID: PMC7338513
- DOI: 10.1038/s41467-020-17292-4
Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia
Abstract
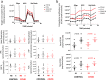
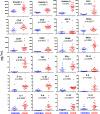
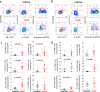
The immune system of patients infected by SARS-CoV-2 is severely impaired. Detailed investigation of T cells and cytokine production in patients affected by COVID-19 pneumonia are urgently required. Here we show that, compared with healthy controls, COVID-19 patients' T cell compartment displays several alterations involving naïve, central memory, effector memory and terminally differentiated cells, as well as regulatory T cells and PD1+CD57+ exhausted T cells. Significant alterations exist also in several lineage-specifying transcription factors and chemokine receptors. Terminally differentiated T cells from patients proliferate less than those from healthy controls, whereas their mitochondria functionality is similar in CD4+ T cells from both groups. Patients display significant increases of proinflammatory or anti-inflammatory cytokines, including T helper type-1 and type-2 cytokines, chemokines and galectins; their lymphocytes produce more tumor necrosis factor (TNF), interferon-γ, interleukin (IL)-2 and IL-17, with the last observation implying that blocking IL-17 could provide a novel therapeutic strategy for COVID-19.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

