Dynamics in protein translation sustaining T cell preparedness
- PMID: 32632289
- PMCID: PMC7610365
- DOI: 10.1038/s41590-020-0714-5
Dynamics in protein translation sustaining T cell preparedness
Abstract
In response to pathogenic threats, naive T cells rapidly transition from a quiescent to an activated state, yet the underlying mechanisms are incompletely understood. Using a pulsed SILAC approach, we investigated the dynamics of mRNA translation kinetics and protein turnover in human naive and activated T cells. Our datasets uncovered that transcription factors maintaining T cell quiescence had constitutively high turnover, which facilitated their depletion following activation. Furthermore, naive T cells maintained a surprisingly large number of idling ribosomes as well as 242 repressed mRNA species and a reservoir of glycolytic enzymes. These components were rapidly engaged following stimulation, promoting an immediate translational and glycolytic switch to ramp up the T cell activation program. Our data elucidate new insights into how T cells maintain a prepared state to mount a rapid immune response, and provide a resource of protein turnover, absolute translation kinetics and protein synthesis rates in T cells ( https://www.immunomics.ch ).
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures


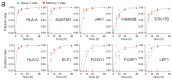




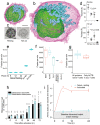

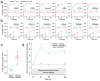
Comment in
-
The active inner life of naive T cells.Nat Immunol. 2020 Aug;21(8):827-828. doi: 10.1038/s41590-020-0726-1. Nat Immunol. 2020. PMID: 32632288 No abstract available.
References
-
- Crabtree GR. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. - PubMed
-
- Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nature reviews Immunology. 2019 - PubMed
-
- Pearce EL, Poffenberger MC, Chang C-H, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454.
-
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nature reviews Immunology. 2005;5:844–852. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

