Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses
- PMID: 32633898
- PMCID: PMC8088761
- DOI: 10.1002/cpcb.110
Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses
Abstract
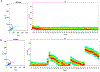
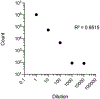
Exosomes are 50- to 150-nm-diameter extracellular vesicles secreted by all mammalian cells except mature red blood cells and contribute to diverse physiological and pathological functions within the body. Many methods have been used to isolate and analyze exosomes, resulting in inconsistencies across experiments and raising questions about how to compare results obtained using different approaches. Questions have also been raised regarding the purity of the various preparations with regard to the sizes and types of vesicles and to the presence of lipoproteins. Thus, investigators often find it challenging to identify the optimal exosome isolation protocol for their experimental needs. Our laboratories have compared ultracentrifugation and commercial precipitation- and column-based exosome isolation kits for exosome preparation. Here, we present protocols for exosome isolation using two of the most commonly used methods, ultracentrifugation and precipitation, followed by downstream analyses. We use NanoSight nanoparticle tracking analysis and flow cytometry (Cytek® ) to determine exosome concentrations and sizes. Imaging flow cytometry can be utilized to both size exosomes and immunophenotype surface markers on exosomes (ImageStream® ). High-performance liquid chromatography followed by nano-flow liquid chromatography-mass spectrometry (LCMS) of the exosome fractions can be used to determine the presence of lipoproteins, with LCMS able to provide a proteomic profile of the exosome preparations. We found that the precipitation method was six times faster and resulted in a ∼2.5-fold higher concentration of exosomes per milliliter compared to ultracentrifugation. Both methods yielded extracellular vesicles in the size range of exosomes, and both preparations included apoproteins. © 2020 Wiley Periodicals LLC. Basic Protocol 1: Pre-analytic fluid collection and processing Basic Protocol 2: Exosome isolation by ultracentrifugation Alternate Protocol 1: Exosome isolation by precipitation Basic Protocol 3: Analysis of exosomes by NanoSight nanoparticle tracking analysis Alternate Protocol 2: Analysis of exosomes by flow cytometry and imaging flow cytometry Basic Protocol 4: Downstream analysis of exosomes using high-performance liquid chromatography Basic Protocol 5: Downstream analysis of the exosome proteome using nano-flow liquid chromatography-mass spectrometry.
Keywords: exosome; isolation; lipoproteins; mass spectrometry; ultracentrifugation.
© 2020 Wiley Periodicals LLC.
Figures


References
-
- Bachurski D, Schuldner M, Nguyen PH, Malz A, Reiners KS, Grenzi PC, … Pogge von Strandmann E (2019). Extracellular vesicle measurements with nanoparticle tracking analysis - An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J Extracell Vesicles, 8(1), 1596016. doi:10.1080/20013078.2019.1596016 - DOI - PMC - PubMed
-
- Feingold KR, & Grunfeld C (2000). Introduction to Lipids and Lipoproteins. In Feingold KR, Anawalt B, Boyce A, Chrousos G, Dungan K, Grossman A, Hershman JM, Kaltsas G, Koch C, Kopp P, Korbonits M, McLachlan R, Morley JE, New M, Perreault L, Purnell J, Rebar R, Singer F, Trence DL, Vinik A, & Wilson DP (Eds.), Endotext. South Dartmouth (MA).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

