The Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade promotes malignant and metastatic progression of osteosarcoma
- PMID: 32634265
- PMCID: PMC7530777
- DOI: 10.1002/1878-0261.12760
The Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade promotes malignant and metastatic progression of osteosarcoma
Retraction in
-
RETRACTION: The Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade promotes malignant and metastatic progression of osteosarcoma.Mol Oncol. 2024 Oct;18(10):2602. doi: 10.1002/1878-0261.13692. Epub 2024 Jul 1. Mol Oncol. 2024. PMID: 38949165 Free PMC article.
Abstract
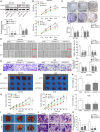
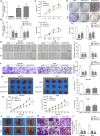
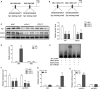
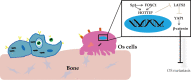
The prognosis for osteosarcoma (OS) is dismal due to the aggressive tumor growth and high incidence of metastasis. The long noncoding RNA human homeobox A transcript at the distal tip (HOTTIP) and the transcription factor forkhead box C1 (FOXC1) present oncogenic activities in OS. Here, we aimed at gaining insights into the underlying mechanisms and their crosstalk. The expression of FOXC1 and HOTTIP in OS tissues or cell lines was examined by real-time PCR (RT-PCR) and western blot. The in vitro effects of FOXC1 or HOTTIP on cell viability, proliferation, migration, invasion, and expression of target genes were examined using MTT, colony-forming assay, wound-healing, Transwell invasion, and western blot, respectively; the in vivo effects were examined using xenograft and experimental metastasis models. Molecular control of HOTTIP on large tumor suppressor 2 (LATS2) or transactivation of FOXC1 or Sp1 on HOTTIP was assessed by combining RNA immunoprecipitation, qRT-PCR, western blot, ChIP, and luciferase assay. Both FOXC1 and HOTTIP were potently up-regulated in OS tissues and cell lines. FOXC1 and HOTTIP essentially maintained viability, proliferation, migration, and invasion of OS cells in vitro and contributed to xenograft growth or lung metastasis in vivo. Mechanistically, HOTTIP recruited enhancer of zeste homolog 2 (EZH2) and lysine-specific demethylase 1 (LSD1) to silence LATS2 and thus activated YAP/β-catenin signaling. Upstream, Sp1 activated FOXC1 and they both directly transactivated HOTTIP. In summary, we showed that the Sp1/FOXC1/HOTTIP/LATS2/YAP/β-catenin cascade presented oncogenic activities in OS cells. Targeting FOXC1 or HOTTIP may therefore prove beneficial for OS treatment.
Keywords: FOXC1; HOTTIP; LATS2; Sp1; osteosarcoma.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Anninga JK, Gelderblom H, Fiocco M, Kroep JR, Taminiau AH, Hogendoorn PC & Egeler RM (2011) Chemotherapeutic adjuvant treatment for osteosarcoma: where do we stand? Eur J Cancer 47, 2431–2445. - PubMed
-
- Gorlick R (2009) Current concepts on the molecular biology of osteosarcoma. Cancer Treat Res 152, 467–478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

