Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis
- PMID: 32634452
- PMCID: PMC8058862
- DOI: 10.1016/j.jaci.2020.06.024
Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis
Abstract
Background: Staphylococcus aureus and Staphylococcus epidermidis are the most abundant bacteria found on the skin of patients with atopic dermatitis (AD). S aureus is known to exacerbate AD, whereas S epidermidis has been considered a beneficial commensal organism.
Objective: In this study, we hypothesized that S epidermidis could promote skin damage in AD by the production of a protease that damages the epidermal barrier.
Methods: The protease activity of S epidermidis isolates was compared with that of other staphylococcal species. The capacity of S epidermidis to degrade the barrier and induce inflammation was examined by using human keratinocyte tissue culture and mouse models. Skin swabs from atopic and healthy adult subjects were analyzed for the presence of S epidermidis genomic DNA and mRNA.
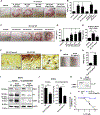
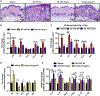
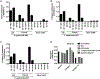
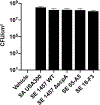
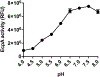
Results: S epidermidis strains were observed to produce strong cysteine protease activity when grown at high density. The enzyme responsible for this activity was identified as EcpA, a cysteine protease under quorum sensing control. EcpA was shown to degrade desmoglein-1 and LL-37 in vitro, disrupt the physical barrier, and induce skin inflammation in mice. The abundance of S epidermidis and expression of ecpA mRNA were increased on the skin of some patients with AD, and this correlated with disease severity. Another commensal skin bacterial species, Staphylococcus hominis, can inhibit EcpA production by S epidermidis.
Conclusion: S epidermidis has commonly been regarded as a beneficial skin microbe, whereas S aureus has been considered deleterious. This study suggests that the overabundance of S epidermidis found on some atopic patients can act similarly to S aureus and damage the skin by expression of a cysteine protease.
Keywords: Atopic dermatitis; Staphylococcus epidermidis; cytokine; dysbiosis; epidermal barrier; inflammation; microbiome; protease; skin.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Disclosure of potential conflict of interest: R. L. Gallo is a cofounder, scientific advisor, and consultant of MatriSys Biosciences and has equity in the company; in addition, he receives income from and has equity in Sente. The rest of the authors declare that they have no relevant conflicts of interest.
Figures


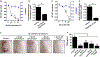
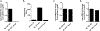
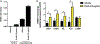
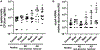
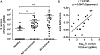

References
-
- Bieber T Atopic dermatitis. N Engl J Med 2008;358:1483–94. - PubMed
-
- Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, Margolis DJ. The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. J Am Acad Dermatol 2008;58:68–73. - PubMed
-
- Carroll CL, Balkrishnan R, Feldman SR, Fleischer AB Jr, Manuel JC. The burden of atopic dermatitis: impact on the patient, family, and society. Pediatr Dermatol 2005;22:192–9. - PubMed
-
- Nutten S Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab 2015;66(Suppl 1):8–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

