Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach
- PMID: 32635935
- PMCID: PMC7339606
- DOI: 10.1186/s12967-020-02439-0
Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach
Abstract
Background: The Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) outbreak originating in Wuhan, China, has raised global health concerns and the pandemic has now been reported on all inhabited continents. Hitherto, no antiviral drug is available to combat this viral outbreak.
Methods: Keeping in mind the urgency of the situation, the current study was designed to devise new strategies for drug discovery and/or repositioning against SARS-CoV-2. In the current study, RNA-dependent RNA polymerase (RdRp), which regulates viral replication, is proposed as a potential therapeutic target to inhibit viral infection.
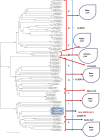
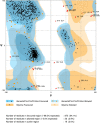
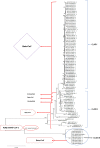
Results: Evolutionary studies of whole-genome sequences of SARS-CoV-2 represent high similarity (> 90%) with other SARS viruses. Targeting the RdRp active sites, ASP760 and ASP761, by antiviral drugs could be a potential therapeutic option for inhibition of coronavirus RdRp, and thus viral replication. Target-based virtual screening and molecular docking results show that the antiviral Galidesivir and its structurally similar compounds have shown promise against SARS-CoV-2.
Conclusions: The anti-polymerase drugs predicted here-CID123624208 and CID11687749-may be considered for in vitro and in vivo clinical trials.
Keywords: Active site; Homology modeling; Molecular Docking; Phylogenetic tree; RdRp; SARS-CoV-2.
Conflict of interest statement
There are no competing interests declared by the authors.
Figures






References
-
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
-
- WHO. Coronavirus Disease 2019 (COVID-19) Situation Report-81. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed 15 Apr 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

