Constructing knowledge graphs and their biomedical applications
- PMID: 32637040
- PMCID: PMC7327409
- DOI: 10.1016/j.csbj.2020.05.017
Constructing knowledge graphs and their biomedical applications
Abstract
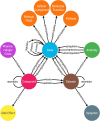
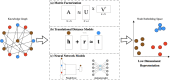
Knowledge graphs can support many biomedical applications. These graphs represent biomedical concepts and relationships in the form of nodes and edges. In this review, we discuss how these graphs are constructed and applied with a particular focus on how machine learning approaches are changing these processes. Biomedical knowledge graphs have often been constructed by integrating databases that were populated by experts via manual curation, but we are now seeing a more robust use of automated systems. A number of techniques are used to represent knowledge graphs, but often machine learning methods are used to construct a low-dimensional representation that can support many different applications. This representation is designed to preserve a knowledge graph's local and/or global structure. Additional machine learning methods can be applied to this representation to make predictions within genomic, pharmaceutical, and clinical domains. We frame our discussion first around knowledge graph construction and then around unifying representational learning techniques and unifying applications. Advances in machine learning for biomedicine are creating new opportunities across many domains, and we note potential avenues for future work with knowledge graphs that appear particularly promising.
Keywords: Lterature review; Machine learning; Natural language processing; Network embeddings; Text mining; knowledge graphs.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

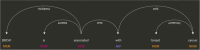

References
-
- Node Classification in Social Networks Smriti Bhagat, Graham Cormode, S. Muthukrishnan Social Network Data Analytics (2011) https://doi.org/fjj48w DOI: 10.1007/978-1-4419-8462-3_5
-
- Network Embedding Based Recommendation Method in Social Networks Yufei Wen, Lei Guo, Zhumin Chen, Jun Ma Companion of the The Web Conference 2018 on The Web Conference 2018 - WWW ’18 (2018) https://doi.org/gf6rtt DOI: 10.1145/3184558.3186904
-
- Open Question Answering with Weakly Supervised Embedding Models Antoine Bordes, Jason Weston, Nicolas Usunier arXiv (2014-04-16) https://arxiv.org/abs/1404.4326v1
-
- Neural Network-based Question Answering over Knowledge Graphs on Word and Character Level Denis Lukovnikov, Asja Fischer, Jens Lehmann, Sören Auer Proceedings of the 26th International Conference on World Wide Web (2017-04-03) https://doi.org/gfv8hp DOI: 10.1145/3038912.3052675
-
- Towards integrative gene prioritization in Alzheimer’s disease. Jang H Lee, Graciela H Gonzalez Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing (2011) https://www.ncbi.nlm.nih.gov/pubmed/21121028 DOI: 10.1142/9789814335058_0002 · PMID: 21121028 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
