This is a preprint.
SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery
- PMID: 32637946
- PMCID: PMC7337376
- DOI: 10.1101/2020.06.29.174623
SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery
Update in
-
SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery.Cell Rep. 2021 May 4;35(5):109055. doi: 10.1016/j.celrep.2021.109055. Epub 2021 Apr 13. Cell Rep. 2021. PMID: 33905739 Free PMC article.
Abstract
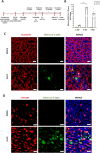
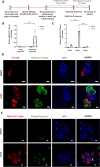
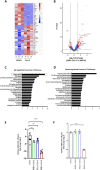
Coronavirus disease 2019 (COVID-19) is the latest respiratory pandemic resulting from zoonotic transmission of severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). Severe symptoms include viral pneumonia secondary to infection and inflammation of the lower respiratory tract, in some cases causing death. We developed primary human lung epithelial infection models to understand responses of proximal and distal lung epithelium to SARS-CoV-2 infection. Differentiated air-liquid interface cultures of proximal airway epithelium and 3D organoid cultures of alveolar epithelium were readily infected by SARS-CoV-2 leading to an epithelial cell-autonomous proinflammatory response. We validated the efficacy of selected candidate COVID-19 drugs confirming that Remdesivir strongly suppressed viral infection/replication. We provide a relevant platform for studying COVID-19 pathobiology and for rapid drug screening against SARS-CoV-2 and future emergent respiratory pathogens.
One sentence summary: A novel infection model of the adult human lung epithelium serves as a platform for COVID-19 studies and drug discovery.
Conflict of interest statement
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
