Expression of NrasQ61R and MYC transgene in germinal center B cells induces a highly malignant multiple myeloma in mice
- PMID: 32640012
- PMCID: PMC7808014
- DOI: 10.1182/blood.2020007156
Expression of NrasQ61R and MYC transgene in germinal center B cells induces a highly malignant multiple myeloma in mice
Abstract
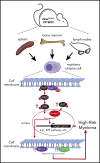
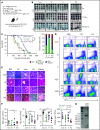
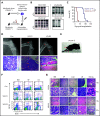
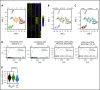
NRAS Q61 mutations are prevalent in advanced/relapsed multiple myeloma (MM) and correlate with poor patient outcomes. Thus, we generated a novel MM model by conditionally activating expression of endogenous NrasQ61R and an MYC transgene in germinal center (GC) B cells (VQ mice). VQ mice developed a highly malignant MM characterized by a high proliferation index, hyperactivation of extracellular signal-regulated kinase and AKT signaling, impaired hematopoiesis, widespread extramedullary disease, bone lesions, kidney abnormalities, preserved programmed cell death protein 1 and T-cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibition motif domain immune-checkpoint pathways, and expression of human high-risk MM gene signatures. VQ MM mice recapitulate most of the biological and clinical features of human advanced/high-risk MM. These MM phenotypes are serially transplantable in syngeneic recipients. Two MM cell lines were also derived to facilitate future genetic manipulations. Combination therapies based on MEK inhibition significantly prolonged the survival of VQ mice with advanced-stage MM. Our study provides a strong rationale to develop MEK inhibition-based therapies for treating advanced/relapsed MM.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



Comment in
-
The winding road toward the myeloma mouse.Blood. 2021 Jan 7;137(1):7-8. doi: 10.1182/blood.2020007942. Blood. 2021. PMID: 33410904 No abstract available.
Similar articles
-
AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies.Cancer Cell. 2008 Feb;13(2):167-80. doi: 10.1016/j.ccr.2008.01.007. Cancer Cell. 2008. PMID: 18242516 Free PMC article.
-
Modeling multiple myeloma by AID-dependent conditional activation of MYC.Cancer Cell. 2008 Feb;13(2):85-7. doi: 10.1016/j.ccr.2008.01.022. Cancer Cell. 2008. PMID: 18242508
-
AID-targeting and hypermutation of non-immunoglobulin genes does not correlate with proximity to immunoglobulin genes in germinal center B cells.PLoS One. 2012;7(6):e39601. doi: 10.1371/journal.pone.0039601. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768095 Free PMC article.
-
Immunocompetent mouse models of multiple myeloma.Semin Hematol. 2025 Feb;62(1):50-57. doi: 10.1053/j.seminhematol.2024.11.003. Epub 2024 Nov 16. Semin Hematol. 2025. PMID: 39674742 Review.
-
Molecular pathogenesis of multiple myeloma: basic and clinical updates.Int J Hematol. 2013 Mar;97(3):313-23. doi: 10.1007/s12185-013-1291-2. Epub 2013 Feb 28. Int J Hematol. 2013. PMID: 23456262 Free PMC article. Review.
Cited by
-
Research Progress on Pyroptosis in Hematological Malignancies.Curr Treat Options Oncol. 2023 Oct;24(10):1439-1450. doi: 10.1007/s11864-023-01119-7. Epub 2023 Aug 28. Curr Treat Options Oncol. 2023. PMID: 37635159 Free PMC article. Review.
-
Molecular characterization stratifies VQ myeloma cells into two clusters with distinct risk signatures and drug responses.Oncogene. 2023 May;42(21):1751-1762. doi: 10.1038/s41388-023-02684-9. Epub 2023 Apr 8. Oncogene. 2023. PMID: 37031341 Free PMC article.
-
Immunocompetent Mouse Models of Multiple Myeloma: Therapeutic Implications.Hematol Oncol Clin North Am. 2024 Apr;38(2):533-546. doi: 10.1016/j.hoc.2023.12.014. Epub 2024 Jan 16. Hematol Oncol Clin North Am. 2024. PMID: 38233233 Free PMC article. Review.
-
Mechanisms of the role of proto-oncogene activation in promoting malignant transformation of mature B cells.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 Jan 28;49(1):113-121. doi: 10.11817/j.issn.1672-7347.2024.230304. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38615172 Free PMC article. Chinese, English.
-
Stromal remodeling regulates dendritic cell abundance and activity in the tumor microenvironment.Cell Rep. 2022 Aug 16;40(7):111201. doi: 10.1016/j.celrep.2022.111201. Cell Rep. 2022. PMID: 35977482 Free PMC article.
References
-
- Kumar SK, Rajkumar V, Kyle RA, et al. . Multiple myeloma. Nat Rev Dis Primers. 2017;3(1):17046. - PubMed
-
- Richter J, Jagannath S. Society of Hematologic Oncology state of the art update and next questions: multiple myeloma. Clin Lymphoma Myeloma Leuk. 2018;18(11):693-702. - PubMed
-
- Papadas A, Asimakopoulos F. Mechanisms of resistance in multiple myeloma. Handb Exp Pharmacol. 2018;249:251-288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

