Characterization of Exome Variants and Their Metabolic Impact in 6,716 American Indians from the Southwest US
- PMID: 32640185
- PMCID: PMC7413855
- DOI: 10.1016/j.ajhg.2020.06.009
Characterization of Exome Variants and Their Metabolic Impact in 6,716 American Indians from the Southwest US
Abstract
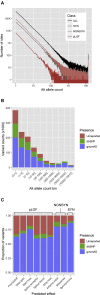
Applying exome sequencing to populations with unique genetic architecture has the potential to reveal novel genes and variants associated with traits and diseases. We sequenced and analyzed the exomes of 6,716 individuals from a Southwestern American Indian (SWAI) population with well-characterized metabolic traits. We found that the SWAI population has distinct allelic architecture compared to populations of European and East Asian ancestry, and there were many predicted loss-of-function (pLOF) and nonsynonymous variants that were highly enriched or private in the SWAI population. We used pLOF and nonsynonymous variants in the SWAI population to evaluate gene-burden associations of candidate genes from European genome-wide association studies (GWASs) for type 2 diabetes, body mass index, and four major plasma lipids. We found 19 significant gene-burden associations for 11 genes, providing additional evidence for prioritizing candidate effector genes of GWAS signals. Interestingly, these associations were mainly driven by pLOF and nonsynonymous variants that are unique or highly enriched in the SWAI population. Particularly, we found four pLOF or nonsynonymous variants in APOB, APOE, PCSK9, and TM6SF2 that are private or enriched in the SWAI population and associated with low-density lipoprotein (LDL) cholesterol levels. Their large estimated effects on LDL cholesterol levels suggest strong impacts on protein function and potential clinical implications of these variants in cardiovascular health. In summary, our study illustrates the utility and potential of exome sequencing in genetically unique populations, such as the SWAI population, to prioritize candidate effector genes within GWAS loci and to find additional variants in known disease genes with potential clinical impact.
Keywords: exome sequencing; gene-burden association; isolated founder population; metabolic traits; rare variant.
Copyright © 2020 American Society of Human Genetics. All rights reserved.
Conflict of interest statement
H.K., B.Y., N.G., A.R.S., and C.V.H. are current or former employees and/or stockholders of Regeneron Genetics Center or Regeneron Pharmaceuticals. H.K. is an employee of Pfizer. N.G. is an employee of RTW Investments. The other authors declare no competing interests.
Figures


References
-
- Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Sequencing Initiative Suomi (SISu) Project Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. - PMC - PubMed
-
- Southam L., Gilly A., Süveges D., Farmaki A.E., Schwartzentruber J., Tachmazidou I., Matchan A., Rayner N.W., Tsafantakis E., Karaleftheri M. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat. Commun. 2017;8:15606. - PMC - PubMed
-
- Rivas M.A., Avila B.E., Koskela J., Huang H., Stevens C., Pirinen M., Haritunians T., Neale B.M., Kurki M., Ganna A., International IBD Genetics Consortium. NIDDK IBD Genetics Consortium. T2D-GENES Consortium Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genet. 2018;14:e1007329. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

