Single-Cell and Population Transcriptomics Reveal Pan-epithelial Remodeling in Type 2-High Asthma
- PMID: 32640237
- PMCID: PMC8046336
- DOI: 10.1016/j.celrep.2020.107872
Single-Cell and Population Transcriptomics Reveal Pan-epithelial Remodeling in Type 2-High Asthma
Abstract
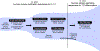
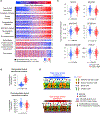
The type 2 cytokine-high asthma endotype (T2H) is characterized by IL-13-driven mucus obstruction of the airways. To further investigate this incompletely understood pathobiology, we characterize IL-13 effects on human airway epithelial cell cultures using single-cell RNA sequencing, finding that IL-13 generates a distinctive transcriptional state for each cell type. Specifically, we discover a mucus secretory program induced by IL-13 in all cell types which converts both mucus and defense secretory cells into a metaplastic state with emergent mucin production and secretion, while leading to ER stress and cell death in ciliated cells. The IL-13-remodeled epithelium secretes a pathologic, mucin-imbalanced, and innate immunity-depleted proteome that arrests mucociliary motion. Signatures of IL-13-induced cellular remodeling are mirrored by transcriptional signatures characteristic of the nasal airway epithelium within T2H versus T2-low asthmatic children. Our results reveal the epithelium-wide scope of T2H asthma and present candidate therapeutic targets for restoring normal epithelial function.
Keywords: GALA; RNA-seq; air-liquid interface; ciliary beat frequency; disease; lung; proteomics; secretome; single cell sequencing; type 2 inflammation.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Beisswenger C, Kandler K, Hess C, Garn H, Felgentreff K, Wegmann M, Renz H, Vogelmeier C, and Bals R (2006). Allergic airway inflammation inhibits pulmonary antibacterial host defense. J. Immunol. 177, 1833–1837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

