NRF2, a Transcription Factor for Stress Response and Beyond
- PMID: 32640524
- PMCID: PMC7369905
- DOI: 10.3390/ijms21134777
NRF2, a Transcription Factor for Stress Response and Beyond
Abstract
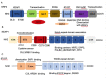
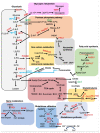
Nuclear factor erythroid 2-related factor 2 (NRF2) is a transcription factor that regulates the cellular defense against toxic and oxidative insults through the expression of genes involved in oxidative stress response and drug detoxification. NRF2 activation renders cells resistant to chemical carcinogens and inflammatory challenges. In addition to antioxidant responses, NRF2 is involved in many other cellular processes, including metabolism and inflammation, and its functions are beyond the originally envisioned. NRF2 activity is tightly regulated through a complex transcriptional and post-translational network that enables it to orchestrate the cell's response and adaptation to various pathological stressors for the homeostasis maintenance. Elevated or decreased NRF2 activity by pharmacological and genetic manipulations of NRF2 activation is associated with many metabolism- or inflammation-related diseases. Emerging evidence shows that NRF2 lies at the center of a complex regulatory network and establishes NRF2 as a truly pleiotropic transcription factor. Here we summarize the complex regulatory network of NRF2 activity and its roles in metabolic reprogramming, unfolded protein response, proteostasis, autophagy, mitochondrial biogenesis, inflammation, and immunity.
Keywords: NRF2; UPR; autophagy; inflammation; metabolism; oxidative stress; proteostasis; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

