Improved reconstruction and comparative analysis of chromosome 12 to rectify Mis-assemblies in Gossypium arboreum
- PMID: 32640982
- PMCID: PMC7346634
- DOI: 10.1186/s12864-020-06814-5
Improved reconstruction and comparative analysis of chromosome 12 to rectify Mis-assemblies in Gossypium arboreum
Abstract
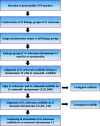
Background: Genome sequencing technologies have been improved at an exponential pace but precise chromosome-scale genome assembly still remains a great challenge. The draft genome of cultivated G. arboreum was sequenced and assembled with shotgun sequencing approach, however, it contains several misassemblies. To address this issue, we generated an improved reassembly of G. arboreum chromosome 12 using genetic mapping and reference-assisted approaches and evaluated this reconstruction by comparing with homologous chromosomes of G. raimondii and G. hirsutum.
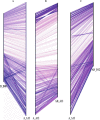
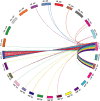
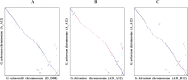
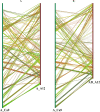
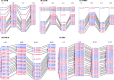
Results: In this study, we generated a high quality assembly of the 94.64 Mb length of G. arboreum chromosome 12 (A_A12) which comprised of 144 scaffolds and contained 3361 protein coding genes. Evaluation of results using syntenic and collinear analysis of reconstructed G. arboreum chromosome A_A12 with its homologous chromosomes of G. raimondii (D_D08) and G. hirsutum (AD_A12 and AD_D12) confirmed the significant improved quality of current reassembly as compared to previous one. We found major misassemblies in previously assembled chromosome 12 (A_Ca9) of G. arboreum particularly in anchoring and orienting of scaffolds into a pseudo-chromosome. Further, homologous chromosomes 12 of G. raimondii (D_D08) and G. arboreum (A_A12) contained almost equal number of transcription factor (TF) related genes, and showed good collinear relationship with each other. As well, a higher rate of gene loss was found in corresponding homologous chromosomes of tetraploid (AD_A12 and AD_D12) than diploid (A_A12 and D_D08) cotton, signifying that gene loss is likely a continuing process in chromosomal evolution of tetraploid cotton.
Conclusion: This study offers a more accurate strategy to correct misassemblies in sequenced draft genomes of cotton which will provide further insights towards its genome organization.
Keywords: Gene loss; Genetic map; Reference-assisted assembly; Syntenic relationship; Transcription factor.
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publication of this paper.
Figures
References
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. - PubMed
-
- Initiative AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. - PubMed
-
- Sasaki T. The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

