Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism
- PMID: 32641298
- PMCID: PMC7370883
- DOI: 10.1101/gr.262774.120
Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism
Abstract
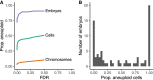
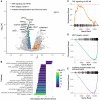
Less than half of human zygotes survive to birth, primarily due to aneuploidies of meiotic or mitotic origin. Mitotic errors generate chromosomal mosaicism, defined by multiple cell lineages with distinct chromosome complements. The incidence and impacts of mosaicism in human embryos remain controversial, with most previous studies based on bulk DNA assays or comparisons of multiple biopsies of few embryonic cells. Single-cell genomic data provide an opportunity to quantify mosaicism on an embryo-wide scale. To this end, we extended an approach to infer aneuploidies based on dosage-associated changes in gene expression by integrating signatures of allelic imbalance. We applied this method to published single-cell RNA sequencing data from 74 human embryos, spanning the morula to blastocyst stages. Our analysis revealed widespread mosaic aneuploidies, with 59 of 74 (80%) embryos harboring at least one putative aneuploid cell (1% FDR). By clustering copy number calls, we reconstructed histories of chromosome segregation, inferring that 55 (74%) embryos possessed mitotic aneuploidies and 23 (31%) embryos possessed meiotic aneuploidies. We found no significant enrichment of aneuploid cells in the trophectoderm compared to the inner cell mass, although we do detect such enrichment in data from later postimplantation stages. Finally, we observed that aneuploid cells up-regulate immune response genes and down-regulate genes involved in proliferation, metabolism, and protein processing, consistent with stress responses documented in other stages and systems. Together, our work provides a high-resolution view of aneuploidy in preimplantation embryos, and supports the conclusion that low-level mosaicism is a common feature of early human development.
© 2020 Starostik et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48. 10.18637/jss.v067.i01 - DOI
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B Methodol 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
