PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity
- PMID: 32641491
- PMCID: PMC7508354
- DOI: 10.1126/scitranslmed.aaz5683
PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity
Abstract
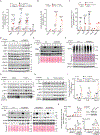
Protein arginine methyltransferase 5 (PRMT5) controls diverse cellular processes and is implicated in cancer development and progression. Here, we report an inverse correlation between PRMT5 function and antitumor immunity. PRMT5 expression was associated with an antitumor immune gene signature in human melanoma tissue. Reducing PRMT5 activity antagonized melanoma growth in immunocompetent but not immunocompromised mice. PRMT5 methylation of IFI16 [interferon-γ (IFN-γ)-inducible protein 16] or its murine homolog IFI204, which are components of the cGAS/STING (stimulator of IFN genes) pathway, attenuated cytosolic DNA-induced IFN and chemokine expression in melanoma cells. PRMT5 also inhibited transcription of the gene encoding NLRC5 (nucleotide-binding oligomerization domain-like receptor family caspase recruitment domain containing 5), a protein that promotes the expression of genes implicated in major histocompatibility complex class I (MHCI) antigen presentation. PRMT5 knockdown augmented IFN and chemokine production and increased MHCI abundance in melanoma. Increased expression of IFI204 and NLRC5 was associated with decreased melanoma growth in murine models, and increased expression of IFI16 and NLRC5 correlated with prolonged survival of patients with melanoma. Combination of pharmacological (GSK3326595) or genetic (shRNA) inhibition of PRMT5 with immune checkpoint therapy limited growth of murine melanoma tumors (B16F10 and YUMM1.7) and enhanced therapeutic efficacy, compared with the effect of either treatment alone. Overall, our findings provide a rationale to test PRMT5 inhibitors in immunotherapy-based clinical trials as a means to enhance an antitumor immune response.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





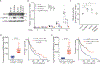

References
-
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbe C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ, Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363, 711–723 (2010). - PMC - PubMed
-
- Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor D, Salama AK, Taylor M, Ott PA, Rollin LM, Horak C, Gagnier P, Wolchok JD, Hodi FS, Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 372, 2006–2017 (2015). - PMC - PubMed
-
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD, Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 373, 23–34 (2015). - PMC - PubMed
-
- Schachter J, Ribas A, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank C, Petrella TM, Hamid O, Zhou H, Ebbinghaus S, Ibrahim N, Robert C, Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 390, 1853–1862 (2017). - PubMed
-
- Spranger S, Bao R, Gajewski TF, Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

