Microbiota modulate sympathetic neurons via a gut-brain circuit
- PMID: 32641826
- PMCID: PMC7367767
- DOI: 10.1038/s41586-020-2474-7
Microbiota modulate sympathetic neurons via a gut-brain circuit
Erratum in
-
Author Correction: Microbiota modulate sympathetic neurons via a gut-brain circuit.Nature. 2020 Sep;585(7823):E2. doi: 10.1038/s41586-020-2657-2. Nature. 2020. PMID: 32814907
Abstract
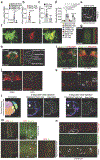
Connections between the gut and brain monitor the intestinal tissue and its microbial and dietary content1, regulating both physiological intestinal functions such as nutrient absorption and motility2,3, and brain-wired feeding behaviour2. It is therefore plausible that circuits exist to detect gut microorganisms and relay this information to areas of the central nervous system that, in turn, regulate gut physiology4. Here we characterize the influence of the microbiota on enteric-associated neurons by combining gnotobiotic mouse models with transcriptomics, circuit-tracing methods and functional manipulations. We find that the gut microbiome modulates gut-extrinsic sympathetic neurons: microbiota depletion leads to increased expression of the neuronal transcription factor cFos, and colonization of germ-free mice with bacteria that produce short-chain fatty acids suppresses cFos expression in the gut sympathetic ganglia. Chemogenetic manipulations, translational profiling and anterograde tracing identify a subset of distal intestine-projecting vagal neurons that are positioned to have an afferent role in microbiota-mediated modulation of gut sympathetic neurons. Retrograde polysynaptic neuronal tracing from the intestinal wall identifies brainstem sensory nuclei that are activated during microbial depletion, as well as efferent sympathetic premotor glutamatergic neurons that regulate gastrointestinal transit. These results reveal microbiota-dependent control of gut-extrinsic sympathetic activation through a gut-brain circuit.
Conflict of interest statement
Competing interests
The authors declare no competing financial interests.
Figures










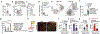
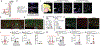

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

