Mitochondrial ubiquinol oxidation is necessary for tumour growth
- PMID: 32641834
- PMCID: PMC7486261
- DOI: 10.1038/s41586-020-2475-6
Mitochondrial ubiquinol oxidation is necessary for tumour growth
Abstract
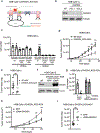
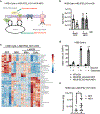
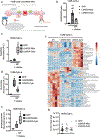
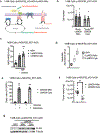
The mitochondrial electron transport chain (ETC) is necessary for tumour growth1-6 and its inhibition has demonstrated anti-tumour efficacy in combination with targeted therapies7-9. Furthermore, human brain and lung tumours display robust glucose oxidation by mitochondria10,11. However, it is unclear why a functional ETC is necessary for tumour growth in vivo. ETC function is coupled to the generation of ATP-that is, oxidative phosphorylation and the production of metabolites by the tricarboxylic acid (TCA) cycle. Mitochondrial complexes I and II donate electrons to ubiquinone, resulting in the generation of ubiquinol and the regeneration of the NAD+ and FAD cofactors, and complex III oxidizes ubiquinol back to ubiquinone, which also serves as an electron acceptor for dihydroorotate dehydrogenase (DHODH)-an enzyme necessary for de novo pyrimidine synthesis. Here we show impaired tumour growth in cancer cells that lack mitochondrial complex III. This phenotype was rescued by ectopic expression of Ciona intestinalis alternative oxidase (AOX)12, which also oxidizes ubiquinol to ubiquinone. Loss of mitochondrial complex I, II or DHODH diminished the tumour growth of AOX-expressing cancer cells deficient in mitochondrial complex III, which highlights the necessity of ubiquinone as an electron acceptor for tumour growth. Cancer cells that lack mitochondrial complex III but can regenerate NAD+ by expression of the NADH oxidase from Lactobacillus brevis (LbNOX)13 targeted to the mitochondria or cytosol were still unable to grow tumours. This suggests that regeneration of NAD+ is not sufficient to drive tumour growth in vivo. Collectively, our findings indicate that tumour growth requires the ETC to oxidize ubiquinol, which is essential to drive the oxidative TCA cycle and DHODH activity.
Conflict of interest statement
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
- I01 CX001777/CX/CSRD VA/United States
- 2T32HL076139-16/CA/NCI NIH HHS/United States
- 5R35CA197532/NH/NIH HHS/United States
- T32 CA009560/CA/NCI NIH HHS/United States
- T32HL076139-13/NH/NIH HHS/United States
- P01 AG049665/AG/NIA NIH HHS/United States
- T32 T32HL076139/NH/NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- F30 CA250236/CA/NCI NIH HHS/United States
- T32 GM008152/GM/NIGMS NIH HHS/United States
- R35 CA197532/CA/NCI NIH HHS/United States
- T32 GM144295/GM/NIGMS NIH HHS/United States
- 5 T32 CA 9560-33/NH/NIH HHS/United States
- T32 HL076139/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

