Fgfr3 mutation disrupts chondrogenesis and bone ossification in zebrafish model mimicking CATSHL syndrome partially via enhanced Wnt/β-catenin signaling
- PMID: 32641982
- PMCID: PMC7330844
- DOI: 10.7150/thno.45286
Fgfr3 mutation disrupts chondrogenesis and bone ossification in zebrafish model mimicking CATSHL syndrome partially via enhanced Wnt/β-catenin signaling
Abstract
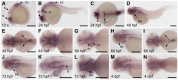
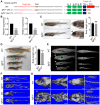
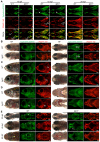
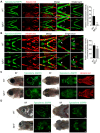
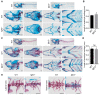
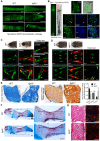
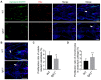
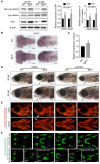
CATSHL syndrome, characterized by camptodactyly, tall stature and hearing loss, is caused by loss-of-function mutations of fibroblast growth factor receptors 3 (FGFR3) gene. Most manifestations of patients with CATSHL syndrome start to develop in the embryonic stage, such as skeletal overgrowth, craniofacial abnormalities, however, the pathogenesis of these phenotypes especially the early maldevelopment remains incompletely understood. Furthermore, there are no effective therapeutic targets for this skeleton dysplasia. Methods: We generated fgfr3 knockout zebrafish by CRISPR/Cas9 technology to study the developmental mechanisms and therapeutic targets of CATSHL syndrome. Several zebrafish transgenic lines labeling osteoblasts and chondrocytes, and live Alizarin red staining were used to analyze the dynamical skeleton development in fgfr3 mutants. Western blotting, whole mount in situ hybridization, Edu labeling based cell proliferation assay and Wnt/β-catenin signaling antagonist were used to explore the potential mechanisms and therapeutic targets. Results: We found that fgfr3 mutant zebrafish, staring from early development stage, showed craniofacial bone malformation with microcephaly and delayed closure of cranial sutures, chondroma-like lesion and abnormal development of auditory sensory organs, partially resembling the clinical manifestations of patients with CATSHL syndrome. Further studies showed that fgfr3 regulates the patterning and shaping of pharyngeal arches and the timely ossification of craniofacial skeleton. The abnormal development of pharyngeal arch cartilage is related to the augmented hypertrophy and disordered arrangement of chondrocytes, while decreased proliferation, differentiation and mineralization of osteoblasts may be involved in the delayed maturation of skull bones. Furthermore, we revealed that deficiency of fgfr3 leads to enhanced IHH signaling and up-regulated canonical Wnt/β-catenin signaling, and pharmacological inhibition of Wnt/β-catenin could partially alleviate the phenotypes of fgfr3 mutants. Conclusions: Our study further reveals some novel phenotypes and underlying developmental mechanism of CATSHL syndrome, which deepens our understanding of the pathogenesis of CATSHL and the role of fgfr3 in skeleton development. Our findings provide evidence that modulation of Wnt/β-catenin activity could be a potential therapy for CATSHL syndrome and related skeleton diseases.
Keywords: CATSHL syndrome; FGFR3; Skeletal development; Wnt/β-catenin; Zebrafish.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Makrythanasis P, Temtamy S, Aglan MS, Otaify GA, Hamamy H, Antonarakis SE. A novel homozygous mutation in FGFR3 causes tall stature, severe lateral tibial deviation, scoliosis, hearing impairment, camptodactyly, and arachnodactyly. Hum Mutat. 2014;35:959–63. - PubMed
-
- Escobar LF, Tucker M, Bamshad M. A second family with CATSHL syndrome: Confirmatory report of another unique FGFR3 syndrome. Am J Med Genet A. 2016;170:1908–11. - PubMed
-
- Ornitz DM, Marie PJ. Fibroblast growth factors in skeletal development. Curr Top Dev Biol. 2019;133:195–234. - PubMed
-
- Du X, Xie Y, Xian CJ, Chen L. Role of FGFs/FGFRs in skeletal development and bone regeneration. J Cell Physiol. 2012;227:3731–43. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

