Improving protein solubility and activity by introducing small peptide tags designed with machine learning models
- PMID: 32642423
- PMCID: PMC7334598
- DOI: 10.1016/j.mec.2020.e00138
Improving protein solubility and activity by introducing small peptide tags designed with machine learning models
Abstract
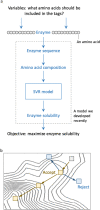
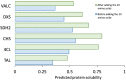
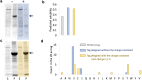
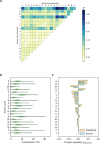
Improving catalytic ability of enzymes is critical to the success of many metabolic engineering projects, but the search space of possible protein mutants is too large to explore exhaustively through experiments. To some extent, highly soluble enzymes tend to exhibit high activity due to their better folding quality. Here, we demonstrate that an optimization algorithm based on a regression model can effectively design short peptide tags to improve solubility of a few model enzymes. Based on the protein sequence information, a support vector regression model we recently developed was used to evaluate protein solubility after small peptide tags were introduced to a target protein. The optimization algorithm guided the sequences of the tags to evolve towards variants that had higher solubility. The optimization results were validated successfully by measuring solubility and activity of the model enzyme with and without the identified tags. The solubility of one protein (tyrosine ammonia lyase) was more than doubled and its activity was improved by 250%. This strategy successfully increased solubility of another two enzymes (aldehyde dehydrogenase and 1-deoxy-D-xylulose-5-phosphate synthase) we tested. The presented optimization methodology thus provides a valuable tool for improving enzyme performance for metabolic engineering and other biotechnology projects.
Keywords: Machine learning; Optimization; Peptide tags; Protein activity; Protein solubility.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Agostini F., Vendruscolo M., Tartaglia G.G. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012;421(2–3):237–241. - PubMed
-
- Bianchi E., Venturini S., Pessi A., Tramontano A., Sollazzo M. High level expression and rational mutagenesis of a designed protein, the minibody: from an insoluble to a soluble molecule. J. Mol. Biol. 1994;236(2):649–659. - PubMed
-
- Bojarski M., Del Testa D., Dworakowski D., Firner B., Flepp B., Goyal P.…Zhang J. 2016. End to End Learning for Self-Driving Cars. arXiv preprint arXiv:1604.07316.
LinkOut - more resources
Full Text Sources
Other Literature Sources

