Proteome-wide data analysis reveals tissue-specific network associated with SARS-CoV-2 infection
- PMID: 32642770
- PMCID: PMC7454804
- DOI: 10.1093/jmcb/mjaa033
Proteome-wide data analysis reveals tissue-specific network associated with SARS-CoV-2 infection
Abstract
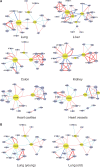
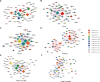
For patients with COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the damages to multiple organs have been clinically observed. Since most of current investigations for virus-host interaction are based on cell level, there is an urgent demand to probe tissue-specific features associated with SARS-CoV-2 infection. Based on collected proteomic datasets from human lung, colon, kidney, liver, and heart, we constructed a virus-receptor network, a virus-interaction network, and a virus-perturbation network. In the tissue-specific networks associated with virus-host crosstalk, both common and different key hubs are revealed in diverse tissues. Ubiquitous hubs in multiple tissues such as BRD4 and RIPK1 would be promising drug targets to rescue multi-organ injury and deal with inflammation. Certain tissue-unique hubs such as REEP5 might mediate specific olfactory dysfunction. The present analysis implies that SARS-CoV-2 could affect multi-targets in diverse host tissues, and the treatment of COVID-19 would be a complex task.
Keywords: SARS-CoV-2; proteome-wide; tissue-specific.
© The Author(s) (2020). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures



References
-
- Bartz F., Kern L., Erz D., et al. (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10, 63–75. - PubMed
-
- Cai G., Cui X., Zhu X., et al. (2020). A hint on the COVID-19 risk: population disparities in gene expression of three receptors of SARS-CoV. Preprints, doi: 10.20944/preprints202002.0408.v1.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

