Genetic diversity and neutral selection in Plasmodium vivax erythrocyte binding protein correlates with patient antigenicity
- PMID: 32645098
- PMCID: PMC7347095
- DOI: 10.1371/journal.pntd.0008202
Genetic diversity and neutral selection in Plasmodium vivax erythrocyte binding protein correlates with patient antigenicity
Abstract
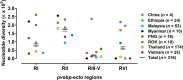
Plasmodium vivax is the most widespread and difficult to treat cause of human malaria. The development of vaccines against the blood stages of P. vivax remains a key objective for the control and elimination of vivax malaria. Erythrocyte binding-like (EBL) protein family members such as Duffy binding protein (PvDBP) are of critical importance to erythrocyte invasion and have been the major target for vivax malaria vaccine development. In this study, we focus on another member of EBL protein family, P. vivax erythrocyte binding protein (PvEBP). PvEBP was first identified in Cambodian (C127) field isolates and has subsequently been showed its preferences for binding reticulocytes which is directly inhibited by antibodies. We analysed PvEBP sequence from 316 vivax clinical isolates from eight countries including China (n = 4), Ethiopia (n = 24), Malaysia (n = 53), Myanmar (n = 10), Papua New Guinea (n = 16), Republic of Korea (n = 10), Thailand (n = 174), and Vietnam (n = 25). PvEBP gene exhibited four different phenotypic clusters based on the insertion/deletion (indels) variation. PvEBP-RII (179-479 aa.) showed highest polymorphism similar to other EBL family proteins in various Plasmodium species. Whereas even though PvEBP-RIII-V (480-690 aa.) was the most conserved domain, that showed strong neutral selection pressure for gene purifying with significant population expansion. Antigenicity of both of PvEBP-RII (16.1%) and PvEBP-RIII-V (21.5%) domains were comparatively lower than other P. vivax antigen which expected antigens associated with merozoite invasion. Total IgG recognition level of PvEBP-RII was stronger than PvEBP-RIII-V domain, whereas total IgG inducing level was stronger in PvEBP-RIII-V domain. These results suggest that PvEBP-RII is mainly recognized by natural IgG for innate protection, whereas PvEBP-RIII-V stimulates IgG production activity by B-cell for acquired immunity. Overall, the low antigenicity of both regions in patients with vivax malaria likely reflects genetic polymorphism for strong positive selection in PvEBP-RII and purifying selection in PvEBP-RIII-V domain. These observations pose challenging questions to the selection of EBP and point out the importance of immune pressure and polymorphism required for inclusion of PvEBP as a vaccine candidate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- World Health Organization. World Malaria Report. 2018.
-
- Kenangalem E, Poespoprodjo JR, Douglas NM, Burdam FH, Gdeumana K, Chalfein F, et al. Malaria morbidity and mortality following introduction of a universal policy of artemisinin-based treatment for malaria in Papua, Indonesia: A longitudinal surveillance study. PLoS medicine. 2019;16(5):e1002815 10.1371/journal.pmed.1002815 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

