Evolution of Mutation Rate in Astronomically Large Phytoplankton Populations
- PMID: 32645145
- PMCID: PMC7486954
- DOI: 10.1093/gbe/evaa131
Evolution of Mutation Rate in Astronomically Large Phytoplankton Populations
Abstract
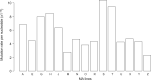
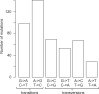
Genetic diversity is expected to be proportional to population size, yet, there is a well-known, but unexplained lack of genetic diversity in large populations-the "Lewontin's paradox." Larger populations are expected to evolve lower mutation rates, which may help to explain this paradox. Here, we test this conjecture by measuring the spontaneous mutation rate in a ubiquitous unicellular marine phytoplankton species Emiliania huxleyi (Haptophyta) that has modest genetic diversity despite an astronomically large population size. Genome sequencing of E. huxleyi mutation accumulation lines revealed 455 mutations, with an unusual GC-biased mutation spectrum. This yielded an estimate of the per site mutation rate µ = 5.55×10-10 (CI 95%: 5.05×10-10 - 6.09×10-10), which corresponds to an effective population size Ne ∼ 2.7×106. Such a modest Ne is surprising for a ubiquitous and abundant species that accounts for up to 10% of global primary productivity in the oceans. Our results indicate that even exceptionally large populations do not evolve mutation rates lower than ∼10-10 per nucleotide per cell division. Consequently, the extreme disparity between modest genetic diversity and astronomically large population size in the plankton species cannot be explained by an unusually low mutation rate.
Keywords: Emiliania huxleyi; Lewontin’s paradox; codon bias; effective population size; mutation accumulation; mutation rate; phytoplankton evolution.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Akashi H. 1997. Codon bias evolution in Drosophila. Population genetics of mutation-selection drift. Gene 205(1–2):269–278. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

