The Global Phosphorylation Landscape of SARS-CoV-2 Infection
- PMID: 32645325
- PMCID: PMC7321036
- DOI: 10.1016/j.cell.2020.06.034
The Global Phosphorylation Landscape of SARS-CoV-2 Infection
Abstract
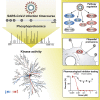
The causative agent of the coronavirus disease 2019 (COVID-19) pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected millions and killed hundreds of thousands of people worldwide, highlighting an urgent need to develop antiviral therapies. Here we present a quantitative mass spectrometry-based phosphoproteomics survey of SARS-CoV-2 infection in Vero E6 cells, revealing dramatic rewiring of phosphorylation on host and viral proteins. SARS-CoV-2 infection promoted casein kinase II (CK2) and p38 MAPK activation, production of diverse cytokines, and shutdown of mitotic kinases, resulting in cell cycle arrest. Infection also stimulated a marked induction of CK2-containing filopodial protrusions possessing budding viral particles. Eighty-seven drugs and compounds were identified by mapping global phosphorylation profiles to dysregulated kinases and pathways. We found pharmacologic inhibition of the p38, CK2, CDK, AXL, and PIKFYVE kinases to possess antiviral efficacy, representing potential COVID-19 therapies.
Keywords: AXL; CDK; MAPK; PIKFYVE; SARS-CoV-2; antiviral; casein kinase II; mass spectrometry; p38; phosphoproteomics.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The Krogan laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. K.M.S. has consulting agreements for the following companies involving cash and/or stock compensation: Black Diamond Therapeutics, BridGene Biosciences, Denali Therapeutics, Dice Molecules, eFFECTOR Therapeutics (zotatifin and tomivosertib), Erasca, Genentech/Roche, Janssen Pharmaceuticals, Kumquat Biosciences, Kura Oncology, Merck, Mitokinin, Petra Pharma, Qulab Inc. Revolution Medicines (WDB002), Type6 Therapeutics, Venthera, and Wellspring Biosciences (Araxes Pharma).
Figures












References
-
- Abraham J., Kelly J., Thibault P., Benchimol S. Post-translational modification of p53 protein in response to ionizing radiation analyzed by mass spectrometry. J. Mol. Biol. 2000;295:853–864. - PubMed
-
- Alfonso R., Lutz T., Rodriguez A., Chavez J.P., Rodriguez P., Gutierrez S., Nieto A. CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell. Microbiol. 2011;13:1894–1906. - PubMed
-
- Ali N., Pruijn G.J., Kenan D.J., Keene J.D., Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000;275:27531–27540. - PubMed
-
- Alvarez D.E., Agaisse H. Casein kinase 2 regulates vaccinia virus actin tail formation. Virology. 2012;423:143–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- R01 GM117189/GM/NIGMS NIH HHS/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- R01 AI122747/AI/NIAID NIH HHS/United States
- F32 CA239333/CA/NCI NIH HHS/United States
- R01 CA244550/CA/NCI NIH HHS/United States
- R35 GM122481/GM/NIGMS NIH HHS/United States
- R01 GM133981/GM/NIGMS NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- T32 CA108462/CA/NCI NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- P50 AI150476/AI/NIAID NIH HHS/United States
- R01 AI143292/AI/NIAID NIH HHS/United States
- F32 CA236347/CA/NCI NIH HHS/United States
- ERC_/European Research Council/International
- R01 AI120694/AI/NIAID NIH HHS/United States
- R35 GM118119/GM/NIGMS NIH HHS/United States
- R01 CA221969/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

