A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS
- PMID: 32645327
- PMCID: PMC7321023
- DOI: 10.1016/j.cell.2020.06.035
A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS
Abstract
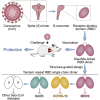
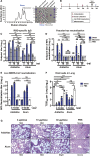
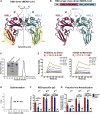
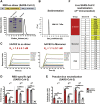
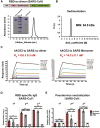
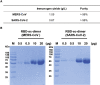
Vaccines are urgently needed to control the ongoing pandemic COVID-19 and previously emerging MERS/SARS caused by coronavirus (CoV) infections. The CoV spike receptor-binding domain (RBD) is an attractive vaccine target but is undermined by limited immunogenicity. We describe a dimeric form of MERS-CoV RBD that overcomes this limitation. The RBD-dimer significantly increased neutralizing antibody (NAb) titers compared to conventional monomeric form and protected mice against MERS-CoV infection. Crystal structure showed RBD-dimer fully exposed dual receptor-binding motifs, the major target for NAbs. Structure-guided design further yielded a stable version of RBD-dimer as a tandem repeat single-chain (RBD-sc-dimer) which retained the vaccine potency. We generalized this strategy to design vaccines against COVID-19 and SARS, achieving 10- to 100-fold enhancement of NAb titers. RBD-sc-dimers in pilot scale production yielded high yields, supporting their scalability for further clinical development. The framework of immunogen design can be universally applied to other beta-CoV vaccines to counter emerging threats.
Keywords: COVID-19; MERS; MERS-CoV; SARS; SARS-CoV; SARS-CoV-2; betacoronavirus; coronavirus; receptor-binding domain (RBD); vaccine.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests L.D., S.L., Y.Y., J.Y., and G.F.G. are listed as inventors on patent applications for MERS-CoV RBD-dimer vaccine. L.D., T.Z., K.X., Y.A., M.L., J.Y., and G.F.G. are listed as inventors on pending patent applications for RBD-sc-dimer-based CoV vaccines. The pending patents for RBD-sc-dimers of MERS-CoV and SARS-CoV-2 have been licensed to Anhui Zhifei Longcom Biopharmaceutical Co. Ltd, China. The other authors declare that they have no competing interests.
Figures



References
-
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

