Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination
- PMID: 32645344
- PMCID: PMC7336130
- DOI: 10.1016/j.lfs.2020.118056
Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination
Abstract
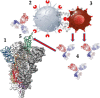
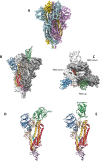
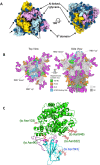
Various human pathogenic viruses employ envelope glycoproteins for host cell receptor recognition and binding, membrane fusion and viral entry. The spike (S) glycoprotein of betacoronavirus SARS-CoV-2 is a homotrimeric class I fusion protein that exists in a metastable conformation for cleavage by host cell proteases furin and TMPRSS2, thereby undergoing substantial structural rearrangement for ACE2 host cell receptor binding and subsequent viral entry by membrane fusion. The S protein is densely decorated with N-linked glycans protruding from the trimer surface that affect S protein folding, processing by host cell proteases and the elicitation of humoral immune response. Deep insight into the sophisticated structure of SARS-CoV-2 S protein may provide a blueprint for vaccination strategies, as reviewed herein.
Keywords: ACE2; Coronavirus SARS-CoV-2; Immune response; Protein structure; Receptor binding domain; Spike protein; Vaccination; Viral fusion protein.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest None.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

