A scoutRNA Is Required for Some Type V CRISPR-Cas Systems
- PMID: 32645367
- PMCID: PMC8196889
- DOI: 10.1016/j.molcel.2020.06.022
A scoutRNA Is Required for Some Type V CRISPR-Cas Systems
Abstract
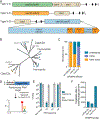
CRISPR-Cas12c/d proteins share limited homology with Cas12a and Cas9 bacterial CRISPR RNA (crRNA)-guided nucleases used widely for genome editing and DNA detection. However, Cas12c (C2c3)- and Cas12d (CasY)-catalyzed DNA cleavage and genome editing activities have not been directly observed. We show here that a short-complementarity untranslated RNA (scoutRNA), together with crRNA, is required for Cas12d-catalyzed DNA cutting. The scoutRNA differs in secondary structure from previously described tracrRNAs used by CRISPR-Cas9 and some Cas12 enzymes, and in Cas12d-containing systems, scoutRNA includes a conserved five-nucleotide sequence that is essential for activity. In addition to supporting crRNA-directed DNA recognition, biochemical and cell-based experiments establish scoutRNA as an essential cofactor for Cas12c-catalyzed pre-crRNA maturation. These results define scoutRNA as a third type of transcript encoded by a subset of CRISPR-Cas genomic loci and explain how Cas12c/d systems avoid requirements for host factors including ribonuclease III for bacterial RNA-mediated adaptive immunity.
Keywords: CRISPR-cas; Candidate Phyla Radiation (CPR) bacteria; Cas12c (C2c3); Cas12d (CasY); RuvC nuclease domain; crRNA; scoutRNA; tracrRNA.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, Algen Biotechnologies, Felix Biosciences, and Inari. J.A.D. is a Director at Johnson & Johnson and has research projects sponsored by Biogen, Pfizer, AppleTree Partners, and Roche. The Regents of the University of California have filed patents related to this work on which D.B., J.F.B., L.B.H., D.P.-E., J.S.C., and J.A.D. are inventors. L.B.H., J.S.C., and J.A.D. are co-founders of Mammoth Biosciences. I.P.W. served as a consultant to Mammoth Biosciences. J.F.B. is a co-founder of Metagenomi.
Figures



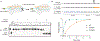
References
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, and Ehrlich SD (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561. - PubMed
-
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, and Banfield JF (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

