Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CLpro targeting repurposed drug candidates
- PMID: 32646487
- PMCID: PMC7344048
- DOI: 10.1186/s12967-020-02448-z
Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CLpro targeting repurposed drug candidates
Abstract
Background: Severe acute respiratory syndrome (SARS) has been initiating pandemics since the beginning of the century. In December 2019, the world was hit again by a devastating SARS episode that has so far infected almost four million individuals worldwide, with over 200,000 fatalities having already occurred by mid-April 2020, and the infection rate continues to grow exponentially. SARS coronavirus 2 (SARS-CoV-2) is a single stranded RNA pathogen which is characterised by a high mutation rate. It is vital to explore the mutagenic capability of the viral genome that enables SARS-CoV-2 to rapidly jump from one host immunity to another and adapt to the genetic pool of local populations.
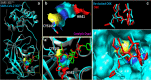
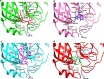
Methods: For this study, we analysed 2301 complete viral sequences reported from SARS-CoV-2 infected patients. SARS-CoV-2 host genomes were collected from The Global Initiative on Sharing All Influenza Data (GISAID) database containing 9 genomes from pangolin-CoV origin and 3 genomes from bat-CoV origin, Wuhan SARS-CoV2 reference genome was collected from GeneBank database. The Multiple sequence alignment tool, Clustal Omega was used for genomic sequence alignment. The viral replicating enzyme, 3-chymotrypsin-like cysteine protease (3CLpro) that plays a key role in its pathogenicity was used to assess its affinity with pharmacological inhibitors and repurposed drugs such as anti-viral flavones, biflavanoids, anti-malarial drugs and vitamin supplements.
Results: Our results demonstrate that bat-CoV shares > 96% similar identity, while pangolin-CoV shares 85.98% identity with Wuhan SARS-CoV-2 genome. This in-depth analysis has identified 12 novel recurrent mutations in South American and African viral genomes out of which 3 were unique in South America, 4 unique in Africa and 5 were present in-patient isolates from both populations. Using state of the art in silico approaches, this study further investigates the interaction of repurposed drugs with the SARS-CoV-2 3CLpro enzyme, which regulates viral replication machinery.
Conclusions: Overall, this study provides insights into the evolving mutations, with implications to understand viral pathogenicity and possible new strategies for repurposing compounds to combat the nCovid-19 pandemic.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

