Clinical Genetic Screening in Adult Patients with Kidney Disease
- PMID: 32646915
- PMCID: PMC7536756
- DOI: 10.2215/CJN.15141219
Clinical Genetic Screening in Adult Patients with Kidney Disease
Abstract
Expanded accessibility of genetic sequencing technologies, such as chromosomal microarray and massively parallel sequencing approaches, is changing the management of hereditary kidney diseases. Genetic causes account for a substantial proportion of pediatric kidney disease cases, and with increased utilization of diagnostic genetic testing in nephrology, they are now also detected at appreciable frequencies in adult populations. Establishing a molecular diagnosis can have many potential benefits for patient care, such as guiding treatment, familial testing, and providing deeper insights on the molecular pathogenesis of kidney diseases. Today, with wider clinical use of genetic testing as part of the diagnostic evaluation, nephrologists have the challenging task of selecting the most suitable genetic test for each patient, and then applying the results into the appropriate clinical contexts. This review is intended to familiarize nephrologists with the various technical, logistical, and ethical considerations accompanying the increasing utilization of genetic testing in nephrology care.
Keywords: CGH array; Chronic; Genetic Testing; High-Throughput Nucleotide Sequencing; Kidney Genomics Series; Patient Care; Renal Insufficiency; Sanger sequencing; array techniques; chronic kidney disease; familial kidney disease; familial nephropathy; genetic renal disease; genetics; genomics; human genetic testing; human genetics; kidney disease; massive parallel sequencing; medical genetics; microarray techniques; nephrology; referral and consultation; translations; whole exome sequencing; whole genome sequencing.
Copyright © 2020 by the American Society of Nephrology.
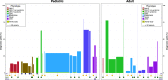
Figures



References
-
- Saran R, Robinson B, Abbott KC, Agodoa LYC, Bragg-Gresham J, Balkrishnan R, Bhave N, Dietrich X, Ding Z, Eggers PW, Gaipov A, Gillen D, Gipson D, Gu H, Guro P, Haggerty D, Han Y, He K, Herman W, Heung M, Hirth RA, Hsiung J-T, Hutton D, Inoue A, Jacobsen SJ, Jin Y, Kalantar-Zadeh K, Kapke A, Kleine C-E, Kovesdy CP, Krueter W, Kurtz V, Li Y, Liu S, Marroquin MV, McCullough K, Molnar MZ, Modi Z, Montez-Rath M, Moradi H, Morgenstern H, Mukhopadhyay P, Nallamothu B, Nguyen DV, Norris KC, O’Hare AM, Obi Y, Park C, Pearson J, Pisoni R, Potukuchi PK, Repeck K, Rhee CM, Schaubel DE, Schrager J, Selewski DT, Shamraj R, Shaw SF, Shi JM, Shieu M, Sim JJ, Soohoo M, Steffick D, Streja E, Sumida K, Kurella Tamura M, Tilea A, Turf M, Wang D, Weng W, Woodside KJ, Wyncott A, Xiang J, Xin X, Yin M, You AS, Zhang X, Zhou H, Shahinian V: US renal data system 2018 annual data report: Epidemiology of kidney disease in the United States. Am J Kidney Dis 73[Suppl 1]: A7–A8, 2019. - PMC - PubMed
-
- Groopman EE, Marasa M, Cameron-Christie S, Petrovski S, Aggarwal VS, Milo-Rasouly H, Li Y, Zhang J, Nestor J, Krithivasan P, Lam WY, Mitrotti A, Piva S, Kil BH, Chatterjee D, Reingold R, Bradbury D, DiVecchia M, Snyder H, Mu X, Mehl K, Balderes O, Fasel DA, Weng C, Radhakrishnan J, Canetta P, Appel GB, Bomback AS, Ahn W, Uy NS, Alam S, Cohen DJ, Crew RJ, Dube GK, Rao MK, Kamalakaran S, Copeland B, Ren Z, Bridgers J, Malone CD, Mebane CM, Dagaonkar N, Fellström BC, Haefliger C, Mohan S, Sanna-Cherchi S, Kiryluk K, Fleckner J, March R, Platt A, Goldstein DB, Gharavi AG: Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 380: 142–151, 2019. - PMC - PubMed
-
- van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, Vivante A, Hwang D-Y, Schulz J, Braun DA, Schmidt JM, Schapiro D, Schneider R, Warejko JK, Daga A, Majmundar AJ, Tan W, Jobst-Schwan T, Hermle T, Widmeier E, Ashraf S, Amar A, Hoogstraaten CA, Hugo H, Kitzler TM, Kause F, Kolvenbach CM, Dai R, Spaneas L, Amann K, Stein DR, Baum MA, Somers MJG, Rodig NM, Ferguson MA, Traum AZ, Daouk GH, Bogdanović R, Stajić N, Soliman NA, Kari JA, El Desoky S, Fathy HM, Milosevic D, Al-Saffar M, Awad HS, Eid LA, Selvin A, Senguttuvan P, Sanna-Cherchi S, Rehm HL, MacArthur DG, Lek M, Laricchia KM, Wilson MW, Mane SM, Lifton RP, Lee RS, Bauer SB, Lu W, Reutter HM, Tasic V, Shril S, Hildebrandt F: Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 29: 2348–2361, 2018. - PMC - PubMed
-
- Connaughton DM, Bukhari S, Conlon P, Cassidy E, O’Toole M, Mohamad M, Flanagan J, Butler T, O’Leary A, Wong L, O’Regan J, Moran S, O’Kelly P, Logan V, Griffin B, Griffin M, Lavin P, Little MA, Conlon P: The Irish kidney gene project--prevalence of family history in patients with kidney disease in Ireland. Nephron 130: 293–301, 2015. - PubMed
-
- van Nimwegen KJM, van Soest RA, Veltman JA, Nelen MR, van der Wilt GJ, Vissers LELM, Grutters JPC: Is the $1000 genome as near as we think? A cost analysis of next-generation sequencing. Clin Chem 62: 1458–1464, 2016. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

