Structure-activity analysis of truncated albumin-binding domains suggests new lead constructs for potential therapeutic delivery
- PMID: 32647013
- PMCID: PMC7443490
- DOI: 10.1074/jbc.RA120.014168
Structure-activity analysis of truncated albumin-binding domains suggests new lead constructs for potential therapeutic delivery
Abstract
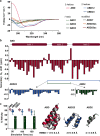
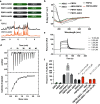
Rapid clearance by renal filtration is a major impediment to the translation of small bioactive biologics into drugs. To extend serum t1/2, a commonly used approach is to attach drug leads to the G-related albumin-binding domain (ABD) to bind albumin and evade clearance. Despite the success of this approach in extending half-lives of a wide range of biologics, it is unclear whether the existing constructs are optimized for binding and size; any improvements along these lines could lead to improved drugs. Characterization of the biophysics of binding of an ABD to albumin in solution could shed light on this question. Here, we examine the binding of an ABD to human serum albumin using isothermal titration calorimetry and assess the structural integrity of the ABD using CD, NMR, and molecular dynamics. A structure-activity analysis of truncations of the ABD suggests that downsized variants could replace the full-length domain. Reducing size could have the benefit of reducing potential immunogenicity problems. We further showed that one of these variants could be used to design a bifunctional molecule with affinity for albumin and a serum protein involved in cholesterol metabolism, PCSK9, demonstrating the potential utility of these fragments in the design of cholesterol-lowering drugs. Future work could extend these in vitro binding studies to other ABD variants to develop therapeutics. Our study presents new understanding of the solution structural and binding properties of ABDs, which has implications for the design of next-generation long-lasting therapeutics.
Keywords: albumin; peptide chemical synthesis; peptide conformation; peptide interaction; peptides.
© 2020 Wang et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

