Visualization of cytoplasmic organelles via in-resin CLEM using an osmium-resistant far-red protein
- PMID: 32647231
- PMCID: PMC7347593
- DOI: 10.1038/s41598-020-68191-z
Visualization of cytoplasmic organelles via in-resin CLEM using an osmium-resistant far-red protein
Abstract
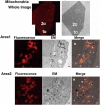
Post-fixation with osmium tetroxide staining and the embedding of Epon are robust and essential treatments that are used to preserve and visualize intracellular membranous structures during electron microscopic analyses. These treatments, however, can significantly diminish the fluorescent intensity of most fluorescent proteins in cells, which creates an obstacle for the in-resin correlative light-electron microscopy (CLEM) of Epon-embedded cells. In this study, we used a far-red fluorescent protein that retains fluorescence after osmium staining and Epon embedding to perform an in-resin CLEM of Epon-embedded samples. The fluorescence of this protein was detected in 100 nm thin sections of the cells in Epon-embedded samples after fixation with 2.5% glutaraldehyde and post-fixation with 1% osmium tetroxide. We performed in-resin CLEM of the mitochondria in Epon-embedded cells using a mitochondria-localized fluorescent protein. Using this protein, we achieved in-resin CLEM of the Golgi apparatus and the endoplasmic reticulum in thin sections of the cells in Epon-embedded samples. To our knowledge, this is the first reported use of a far-red fluorescent protein retains its fluorescence after osmium staining and Epon-embedding, and it represents the first achievement of in-resin CLEM of both the Golgi apparatus and the endoplasmic reticulum in Epon-embedded samples.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

