Patient-specific Alzheimer-like pathology in trisomy 21 cerebral organoids reveals BACE2 as a gene dose-sensitive AD suppressor in human brain
- PMID: 32647257
- PMCID: PMC8190957
- DOI: 10.1038/s41380-020-0806-5
Patient-specific Alzheimer-like pathology in trisomy 21 cerebral organoids reveals BACE2 as a gene dose-sensitive AD suppressor in human brain
Erratum in
-
Correction: Patient-specific Alzheimer-like pathology in trisomy 21 cerebral organoids reveals BACE2 as a gene dose-sensitive AD suppressor in human brain.Mol Psychiatry. 2021 Oct;26(10):5789. doi: 10.1038/s41380-021-01206-x. Mol Psychiatry. 2021. PMID: 34272490 Free PMC article. No abstract available.
Abstract
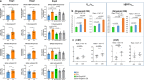
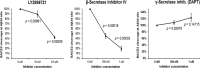
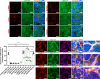
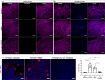
A population of more than six million people worldwide at high risk of Alzheimer's disease (AD) are those with Down Syndrome (DS, caused by trisomy 21 (T21)), 70% of whom develop dementia during lifetime, caused by an extra copy of β-amyloid-(Aβ)-precursor-protein gene. We report AD-like pathology in cerebral organoids grown in vitro from non-invasively sampled strands of hair from 71% of DS donors. The pathology consisted of extracellular diffuse and fibrillar Aβ deposits, hyperphosphorylated/pathologically conformed Tau, and premature neuronal loss. Presence/absence of AD-like pathology was donor-specific (reproducible between individual organoids/iPSC lines/experiments). Pathology could be triggered in pathology-negative T21 organoids by CRISPR/Cas9-mediated elimination of the third copy of chromosome 21 gene BACE2, but prevented by combined chemical β and γ-secretase inhibition. We found that T21 organoids secrete increased proportions of Aβ-preventing (Aβ1-19) and Aβ-degradation products (Aβ1-20 and Aβ1-34). We show these profiles mirror in cerebrospinal fluid of people with DS. We demonstrate that this protective mechanism is mediated by BACE2-trisomy and cross-inhibited by clinically trialled BACE1 inhibitors. Combined, our data prove the physiological role of BACE2 as a dose-sensitive AD-suppressor gene, potentially explaining the dementia delay in ~30% of people with DS. We also show that DS cerebral organoids could be explored as pre-morbid AD-risk population detector and a system for hypothesis-free drug screens as well as identification of natural suppressor genes for neurodegenerative diseases.
© 2020. The Author(s).
Conflict of interest statement
HZ has served at scientific advisory boards for CogRx, Roche Diagnostics, Samumed, and Wave and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg (all unrelated to the submitted work). JEM chairs the MNDA Research Advisory Board and is a director of Biomoti, a cancer drug-delivery company.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

