SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects
- PMID: 32647346
- PMCID: PMC7610980
- DOI: 10.1038/s41594-020-0468-7
SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects
Erratum in
-
Author Correction: SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.Nat Struct Mol Biol. 2020 Oct;27(10):1001. doi: 10.1038/s41594-020-0509-2. Nat Struct Mol Biol. 2020. PMID: 32848232 Free PMC article.
Abstract
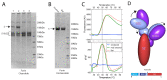
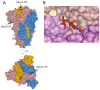
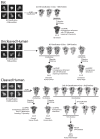
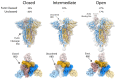
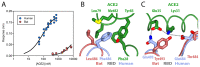
SARS-CoV-2 is thought to have emerged from bats, possibly via a secondary host. Here, we investigate the relationship of spike (S) glycoprotein from SARS-CoV-2 with the S protein of a closely related bat virus, RaTG13. We determined cryo-EM structures for RaTG13 S and for both furin-cleaved and uncleaved SARS-CoV-2 S; we compared these with recently reported structures for uncleaved SARS-CoV-2 S. We also biochemically characterized their relative stabilities and affinities for the SARS-CoV-2 receptor ACE2. Although the overall structures of human and bat virus S proteins are similar, there are key differences in their properties, including a more stable precleavage form of human S and about 1,000-fold tighter binding of SARS-CoV-2 to human receptor. These observations suggest that cleavage at the furin-cleavage site decreases the overall stability of SARS-CoV-2 S and facilitates the adoption of the open conformation that is required for S to bind to the ACE2 receptor.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lai M, Perlman S, Anderson L. Coronaviridae. Fields Virology. 2013:1305–1336.
-
- Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science (80- ) 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

