The effects of mutant Ras proteins on the cell signalome
- PMID: 32648136
- PMCID: PMC7680337
- DOI: 10.1007/s10555-020-09912-8
The effects of mutant Ras proteins on the cell signalome
Abstract
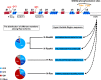
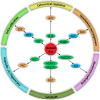
The genetic alterations in cancer cells are tightly linked to signaling pathway dysregulation. Ras is a key molecule that controls several tumorigenesis-related processes, and mutations in RAS genes often lead to unbiased intensification of signaling networks that fuel cancer progression. In this article, we review recent studies that describe mutant Ras-regulated signaling routes and their cross-talk. In addition to the two main Ras-driven signaling pathways, i.e., the RAF/MEK/ERK and PI3K/AKT/mTOR pathways, we have also collected emerging data showing the importance of Ras in other signaling pathways, including the RAC/PAK, RalGDS/Ral, and PKC/PLC signaling pathways. Moreover, microRNA-regulated Ras-associated signaling pathways are also discussed to highlight the importance of Ras regulation in cancer. Finally, emerging data show that the signal alterations in specific cell types, such as cancer stem cells, could promote cancer development. Therefore, we also cover the up-to-date findings related to Ras-regulated signal transduction in cancer stem cells.
Keywords: Mutant Ras protein; Phosphorylation; Signal transduction; Tumorigenesis.
Conflict of interest statement
The authors declare that they have no competing interests
Figures

References
-
- Xu J, Pfarr N, Endris V, Mai EK, Md Hanafiah NH, Lehners N, Penzel R, Weichert W, Ho AD, Schirmacher P, Goldschmidt H, Andrulis M, Raab MS. Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation. Oncogenesis. 2017;6(5):e337–e337. doi: 10.1038/oncsis.2017.36. - DOI - PMC - PubMed
-
- Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D, Halilovic E, Wilson M, Huberman K, Ricarte Filho JC, Persaud Y, Levine DA, Fagin JA, Jhanwar SC, Mariadason JM, Lash A, Ladanyi M, Saltz LB, Heguy A, Paty PB, Solit DB. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Research. 2010;70(14):5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. - DOI - PMC - PubMed
-
- Leiser D, Medová M, Mikami K, Nisa L, Stroka D, Blaukat A, Bladt F, Aebersold DM, Zimmer Y. KRAS and HRAS mutations confer resistance to MET targeting in preclinical models of MET-expressing tumor cells. Molecular Oncology. 2015;9(7):1434–1446. doi: 10.1016/j.molonc.2015.04.001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

