Structural basis for RIFIN-mediated activation of LILRB1 in malaria
- PMID: 32650338
- PMCID: PMC7116854
- DOI: 10.1038/s41586-020-2530-3
Structural basis for RIFIN-mediated activation of LILRB1 in malaria
Abstract
The Plasmodium species that cause malaria are obligate intracellular parasites, and disease symptoms occur when these parasites replicate in human blood. Despite the risk of immune detection, the parasite delivers proteins that bind to host receptors on the cell surfaces of infected erythrocytes. In the causative parasite of the most deadly form of malaria in humans, Plasmodium falciparum, RIFINs form the largest family of surface proteins displayed by erythrocytes1. Some RIFINs can bind to inhibitory immune receptors, and these RIFINs act as targets for unusual antibodies that contain a LAIR1 ectodomain2-4 or as ligands for LILRB15. RIFINs stimulate the activation of and signalling by LILRB15, which could potentially lead to the dampening of human immune responses. Here, to understand how RIFINs activate LILRB1-mediated signalling, we determine the structure of a RIFIN bound to LILRB1. We show that this RIFIN mimics the natural activating ligand of LILRB1, MHC class I, in its LILRB1-binding mode. A single mutation in the RIFIN disrupts the complex, blocks LILRB1 binding of all tested RIFINs and abolishes signalling in a reporter assay. In a supported lipid bilayer system, which mimics the activation of natural killer (NK) cells by antibody-dependent cell-mediated cytotoxicity, both RIFIN and MHC are recruited to the immunological synapse of NK cells and reduce the activation of NK cells, as measured by the mobilization of perforin. Therefore, LILRB1-binding RIFINs mimic the binding mode of the natural ligand of LILRB1 and suppress the function of NK cells.
Conflict of interest statement
Figures
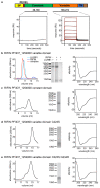
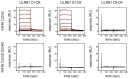



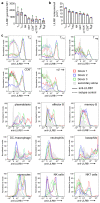

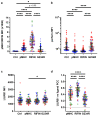


Comment in
-
RIFINing Plasmodium-NK Cell Interaction.Trends Parasitol. 2020 Oct;36(10):802-804. doi: 10.1016/j.pt.2020.07.017. Epub 2020 Aug 12. Trends Parasitol. 2020. PMID: 32800428
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

