Escher-Trace: a web application for pathway-based visualization of stable isotope tracing data
- PMID: 32650717
- PMCID: PMC7350651
- DOI: 10.1186/s12859-020-03632-0
Escher-Trace: a web application for pathway-based visualization of stable isotope tracing data
Abstract
Background: Stable isotope tracing has become an invaluable tool for probing the metabolism of biological systems. However, data analysis and visualization from metabolic tracing studies often involve multiple software packages and lack pathway architecture. A deep understanding of the metabolic contexts from such datasets is required for biological interpretation. Currently, there is no single software package that allows researchers to analyze and integrate stable isotope tracing data into annotated or custom-built metabolic networks.
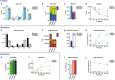
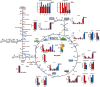
Results: We built a standalone web-based software, Escher-Trace, for analyzing tracing data and communicating results. Escher-Trace allows users to upload baseline corrected mass spectrometer (MS) tracing data and correct for natural isotope abundance, generate publication quality graphs of metabolite labeling, and present data in the context of annotated metabolic pathways. Here we provide a detailed walk-through of how to incorporate and visualize 13C metabolic tracing data into the Escher-Trace platform.
Conclusions: Escher-Trace is an open-source software for analysis and interpretation of stable isotope tracing data and is available at https://escher-trace.github.io/ .
Keywords: Escher; Metabolism; Stable isotope tracing; Visualization; Web application.
Conflict of interest statement
The authors declare that they have no conflict of interest with the contents of this article.
Figures