Quantitative planar array screen of 1000 proteins uncovers novel urinary protein biomarkers of lupus nephritis
- PMID: 32651195
- PMCID: PMC7839323
- DOI: 10.1136/annrheumdis-2019-216312
Quantitative planar array screen of 1000 proteins uncovers novel urinary protein biomarkers of lupus nephritis
Abstract
Objective: The goal of these studies is to discover novel urinary biomarkers of lupus nephritis (LN).
Methods: Urine from systemic lupus erythematosus (SLE) patients was interrogated for 1000 proteins using a novel, quantitative planar protein microarray. Hits were validated in an independent SLE cohort with inactive, active non-renal (ANR) and active renal (AR) patients, in a cohort with concurrent renal biopsies, and in a longitudinal cohort. Single-cell renal RNA sequencing data from LN kidneys were examined to deduce the cellular origin of each biomarker.
Results: Screening of 1000 proteins revealed 64 proteins to be significantly elevated in SLE urine, of which 17 were ELISA validated in independent cohorts. Urine Angptl4 (area under the curve (AUC)=0.96), L-selectin (AUC=0.86), TPP1 (AUC=0.84), transforming growth factor-β1 (TGFβ1) (AUC=0.78), thrombospondin-1 (AUC=0.73), FOLR2 (AUC=0.72), platelet-derived growth factor receptor-β (AUC=0.67) and PRX2 (AUC=0.65) distinguished AR from ANR SLE, outperforming anti-dsDNA, C3 and C4, in terms of specificity, sensitivity and positive predictive value. In multivariate regression analysis, urine Angptl4, L-selectin, TPP1 and TGFβ1 were highly associated with disease activity, even after correction for demographic variables. In SLE patients with serial follow-up, urine L-selectin (followed by urine Angptl4 and TGFβ1) were best at tracking concurrent or pending disease flares. Importantly, several proteins elevated in LN urine were also expressed within the kidneys in LN, either within resident renal cells or infiltrating immune cells, based on single-cell RNA sequencing analysis.
Conclusion: Unbiased planar array screening of 1000 proteins has led to the discovery of urine Angptl4, L-selectin and TGFβ1 as potential biomarker candidates for tracking disease activity in LN.
Keywords: autoimmunity; cytokines; lupus nephritis; systemic lupus erythematosus.
© Author(s) (or their employer(s)) 2020. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

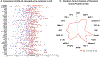

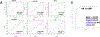



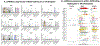
References
-
- Pisetsky DS. Anti-DNA antibodies — quintessential biomarkers of SLE. Nature Reviews Rheumatology 2016;12:102–10. - PubMed
-
- Soliman S, Chandra Mohan. Lupus nephritis biomarkers. Clinical Immunology 2017;185:10–20. - PubMed
-
- Rovin BH, Song H, Birmingham DJ, Hebert LA, Yu CY, Nagaraja HN. Urine Chemokines as Biomarkers of Human Systemic Lupus Erythematosus Activity. JASN 2005;16:467–73. - PubMed
-
- Schwartz N, Michaelson JS, Putterman C. Lipocalin-2, TWEAK, and Other Cytokines as Urinary Biomarkers for Lupus Nephritis. Annals of the New York Academy of Sciences 2007;1109:265–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

