Pharmacologic induction of innate immune signaling directly drives homologous recombination deficiency
- PMID: 32651270
- PMCID: PMC7395437
- DOI: 10.1073/pnas.2003499117
Pharmacologic induction of innate immune signaling directly drives homologous recombination deficiency
Abstract
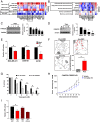
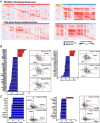
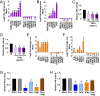
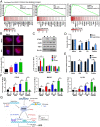
Poly(ADP ribose) polymerase inhibitors (PARPi) have efficacy in triple negative breast (TNBC) and ovarian cancers (OCs) harboring BRCA mutations, generating homologous recombination deficiencies (HRDs). DNA methyltransferase inhibitors (DNMTi) increase PARP trapping and reprogram the DNA damage response to generate HRD, sensitizing BRCA-proficient cancers to PARPi. We now define the mechanisms through which HRD is induced in BRCA-proficient TNBC and OC. DNMTi in combination with PARPi up-regulate broad innate immune and inflammasome-like signaling events, driven in part by stimulator of interferon genes (STING), to unexpectedly directly generate HRD. This inverse relationship between inflammation and DNA repair is critical, not only for the induced phenotype, but also appears as a widespread occurrence in The Cancer Genome Atlas datasets and cancer subtypes. These discerned interactions between inflammation signaling and DNA repair mechanisms now elucidate how epigenetic therapy enhances PARPi efficacy in the setting of BRCA-proficient cancer. This paradigm will be tested in a phase I/II TNBC clinical trial.
Keywords: DNA methyltransferase inhibitors; Fanconi anemia; homologous recombination deficiency; poly(ADP-ribose) polymerase inhibitors; stimulator of interferon signaling.
Conflict of interest statement
Competing interest statement: F.V.R. and S.B.B. are co-inventors on US Provisional Patent Application Number 61/929,680 for the concept of the combinatorial therapy.
Figures


References
-
- De Vos M., Schreiber V., Dantzer F., The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol. 84, 137–146 (2012). - PubMed
-
- Audebert M., Salles B., Calsou P., Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 (2004). - PubMed
-
- Gibson B. A., Kraus W. L., New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 (2012). - PubMed
-
- Chevanne M. et al. ., Inhibition of PARP activity by PJ-34 leads to growth impairment and cell death associated with aberrant mitotic pattern and nucleolar actin accumulation in M14 melanoma cell line. J. Cell. Physiol. 222, 401–410 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

