Raman image-activated cell sorting
- PMID: 32651381
- PMCID: PMC7351993
- DOI: 10.1038/s41467-020-17285-3
Raman image-activated cell sorting
Abstract
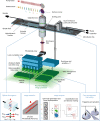
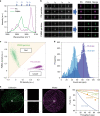
The advent of image-activated cell sorting and imaging-based cell picking has advanced our knowledge and exploitation of biological systems in the last decade. Unfortunately, they generally rely on fluorescent labeling for cellular phenotyping, an indirect measure of the molecular landscape in the cell, which has critical limitations. Here we demonstrate Raman image-activated cell sorting by directly probing chemically specific intracellular molecular vibrations via ultrafast multicolor stimulated Raman scattering (SRS) microscopy for cellular phenotyping. Specifically, the technology enables real-time SRS-image-based sorting of single live cells with a throughput of up to ~100 events per second without the need for fluorescent labeling. To show the broad utility of the technology, we show its applicability to diverse cell types and sizes. The technology is highly versatile and holds promise for numerous applications that are previously difficult or undesirable with fluorescence-based technologies.
Conflict of interest statement
Y.Su., Y.Oz., and K.G. are inventors of a patent application covering SRS imaging with a wavelength-switched laser. S.S., F.A., and T.Hay. are inventors of a patent application covering the dual-membrane push-pull cell sorter. N.N., T.Su., and K.G. are shareholders of CYBO. The remaining authors declare no competing interests.
Figures


References
-
- Nitta N, et al. Intelligent image-activated cell sorting. Cell. 2018;175:266–276. - PubMed
-
- Isozaki A, et al. A practical guide to intelligent image-activated cell sorting. Nat. Protoc. 2019;14:2370–2415. - PubMed
-
- Jin A, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat. Med. 2009;15:1088–1092. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

