Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation
- PMID: 32651390
- PMCID: PMC7351774
- DOI: 10.1038/s41467-020-17327-w
Within-host microevolution of Streptococcus pneumoniae is rapid and adaptive during natural colonisation
Abstract
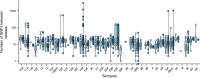
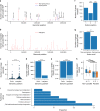
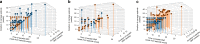
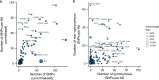
Genomic evolution, transmission and pathogenesis of Streptococcus pneumoniae, an opportunistic human-adapted pathogen, is driven principally by nasopharyngeal carriage. However, little is known about genomic changes during natural colonisation. Here, we use whole-genome sequencing to investigate within-host microevolution of naturally carried pneumococci in ninety-eight infants intensively sampled sequentially from birth until twelve months in a high-carriage African setting. We show that neutral evolution and nucleotide substitution rates up to forty-fold faster than observed over longer timescales in S. pneumoniae and other bacteria drives high within-host pneumococcal genetic diversity. Highly divergent co-existing strain variants emerge during colonisation episodes through real-time intra-host homologous recombination while the rest are co-transmitted or acquired independently during multiple colonisation episodes. Genic and intergenic parallel evolution occur particularly in antibiotic resistance, immune evasion and epithelial adhesion genes. Our findings suggest that within-host microevolution is rapid and adaptive during natural colonisation.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Brueggemann AB, et al. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 2003;187:1424–1432. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

