Group structure and kinship in beluga whale societies
- PMID: 32651398
- PMCID: PMC7351962
- DOI: 10.1038/s41598-020-67314-w
Group structure and kinship in beluga whale societies
Abstract
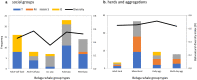
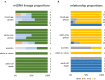
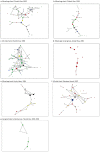
Evolutionary explanations for mammalian sociality typically center on inclusive-fitness benefits of associating and cooperating with close kin, or close maternal kin as in some whale societies, including killer and sperm whales. Their matrilineal structure has strongly influenced the thinking about social structure in less well-studied cetaceans, including beluga whales. In a cross-sectional study of group structure and kinship we found that belugas formed a limited number of distinct group types, consistently observed across populations and habitats. Certain behaviours were associated with group type, but group membership was often dynamic. MtDNA-microsatellite profiling combined with relatedness and network analysis revealed, contrary to predictions, that most social groupings were not predominantly organized around close maternal relatives. They comprised both kin and non-kin, many group members were paternal rather than maternal relatives, and unrelated adult males often traveled together. The evolutionary mechanisms that shape beluga societies are likely complex; fitness benefits may be achieved through reciprocity, mutualism and kin selection. At the largest scales these societies are communities comprising all ages and both sexes where multiple social learning pathways involving kin and non-kin can foster the emergence of cultures. We explore the implications of these findings for species management and the evolution of menopause.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Hamilton WD. The genetical evolution of social behavior I. J. Theor. Biol. 1964;7:1–6. - PubMed
-
- Trivers RL. The evolution of reciprocal altruism. Q. Rev. Biol. 1971;46:35–57.
-
- Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. - PubMed
-
- Maynard Smith J. Game theory and the evolution of cooperation. In: Bendall DS, editor. Evolution from Molecules to Men. Cambridge: Cambridge University Press; 1983. pp. 445–456.
-
- Clutton-Brock T. Cooperation between non-kin in animal societies. Nature. 2009;462:51–57. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

