Current trends in gene recovery mediated by the CRISPR-Cas system
- PMID: 32651459
- PMCID: PMC8080666
- DOI: 10.1038/s12276-020-0466-1
Current trends in gene recovery mediated by the CRISPR-Cas system
Abstract
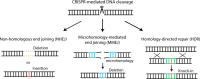
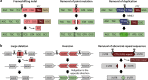
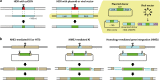
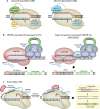
The CRISPR-Cas system has undoubtedly revolutionized the genome editing field, enabling targeted gene disruption, regulation, and recovery in a guide RNA-specific manner. In this review, we focus on currently available gene recovery strategies that use CRISPR nucleases, particularly for the treatment of genetic disorders. Through the action of DNA repair mechanisms, CRISPR-mediated DNA cleavage at a genomic target can shift the reading frame to correct abnormal frameshifts, whereas DNA cleavage at two sites, which can induce large deletions or inversions, can correct structural abnormalities in DNA. Homology-mediated or homology-independent gene recovery strategies that require donor DNAs have been developed and widely applied to precisely correct mutated sequences in genes of interest. In contrast to the DNA cleavage-mediated gene correction methods listed above, base-editing tools enable base conversion in the absence of donor DNAs. In addition, CRISPR-associated transposases have been harnessed to generate a targeted knockin, and prime editors have been developed to edit tens of nucleotides in cells. Here, we introduce currently developed gene recovery strategies and discuss the pros and cons of each.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

