Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein
- PMID: 32651581
- PMCID: PMC8194108
- DOI: 10.1038/s41591-020-0998-x
Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein
Abstract
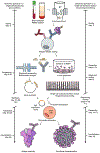
Antibodies are a principal determinant of immunity for most RNA viruses and have promise to reduce infection or disease during major epidemics. The novel coronavirus SARS-CoV-2 has caused a global pandemic with millions of infections and hundreds of thousands of deaths to date1,2. In response, we used a rapid antibody discovery platform to isolate hundreds of human monoclonal antibodies (mAbs) against the SARS-CoV-2 spike (S) protein. We stratify these mAbs into five major classes on the basis of their reactivity to subdomains of S protein as well as their cross-reactivity to SARS-CoV. Many of these mAbs inhibit infection of authentic SARS-CoV-2 virus, with most neutralizing mAbs recognizing the receptor-binding domain (RBD) of S. This work defines sites of vulnerability on SARS-CoV-2 S and demonstrates the speed and robustness of advanced antibody discovery platforms.
Figures









Update of
-
Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein.bioRxiv [Preprint]. 2020 May 13:2020.05.12.091462. doi: 10.1101/2020.05.12.091462. bioRxiv. 2020. Update in: Nat Med. 2020 Sep;26(9):1422-1427. doi: 10.1038/s41591-020-0998-x. PMID: 32511414 Free PMC article. Updated. Preprint.
References
Methods References
-
- Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). - PubMed
-
- Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

