Tumor organoids to study gastroesophageal cancer: a primer
- PMID: 32652008
- PMCID: PMC7683018
- DOI: 10.1093/jmcb/mjaa035
Tumor organoids to study gastroesophageal cancer: a primer
Abstract
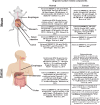
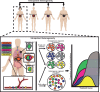
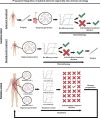
Gastroesophageal cancers are leading causes of cancer death. Our attempts at adopting molecularly based treatment approaches have been slow and ineffective even though we begin to identify specific targetable gene mutations and pathways. It is clear that we should no longer treat all gastroesophageal cancers as a homogeneous disease, which is what we do when we use non-specific chemotherapy. However, we currently cannot monitor successful gene/pathway targeting, nor understand how/when tumors develop resistance, nor predict which patients will derive maximal benefit. To improve outcomes, we must precisely detail the heterogeneity of these tumors to then individualize cancer therapy as well as develop novel avenues to study and predict treatment effects in individual patients. To this end, patient-derived organoids, in which tumor cells from individual patients are grown in a Petri dish, are a new versatile system that allows for timely expandability, detailed molecular characterization, and genetic manipulation with the promise of enabling predictive assessment of treatment response. In this review, we will explore the development and basic techniques for organoid generation, and discuss the current and potential future applications of this exciting technology to study the basic science of carcinogenesis and to predict/guide cancer patient care in the clinics.
Keywords: cancer evolution; cancer model; personalized medicine; precision oncology; targeted therapy; tumor heterogeneity.
© The Author(s) (2020). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures

References
-
- Bang Y.J., Van Cutsem E., Feyereislova A., et al. (2010). Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697. - PubMed
-
- Barker N., Huch M., Kujala P., et al. (2010). Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36. - PubMed
-
- Barker N., Ridgway R.A., van Es J.H., et al. (2009). Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611. - PubMed
-
- Barker N., van Es J.H., Kuipers J., et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

