Scleral HIF-1α is a prominent regulatory candidate for genetic and environmental interactions in human myopia pathogenesis
- PMID: 32652319
- PMCID: PMC7348000
- DOI: 10.1016/j.ebiom.2020.102878
Scleral HIF-1α is a prominent regulatory candidate for genetic and environmental interactions in human myopia pathogenesis
Abstract
Background: Myopia is a good model for understanding the interaction between genetics and environmental stimuli. Here we dissect the biological processes affecting myopia progression.
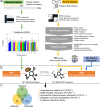
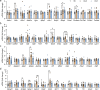
Methods: Human Genetic Analyses: (1) gene set analysis (GSA) of new genome wide association study (GWAS) data for 593 individuals with high myopia (refraction ≤ -6 diopters [D]); (2) over-representation analysis (ORA) of 196 genes with de novo mutations, identified by whole genome sequencing of 45 high-myopia trio families, and (3) ORA of 284 previously reported myopia risk genes. Contributions of the enriched signaling pathways in mediating the genetic and environmental interactions during myopia development were investigated in vivo and in vitro.
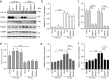
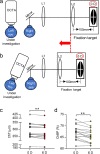
Results: All three genetic analyses showed significant enrichment of four KEGG signaling pathways, including amphetamine addiction, extracellular matrix (ECM) receptor interaction, neuroactive ligand-receptor interaction, and regulation of actin cytoskeleton pathways. In individuals with extremely high myopia (refraction ≤ -10 D), the GSA of GWAS data revealed significant enrichment of the HIF-1α signaling pathway. Using human scleral fibroblasts, silencing the key nodal genes within protein-protein interaction networks for the enriched pathways antagonized the hypoxia-induced increase in myofibroblast transdifferentiation. In mice, scleral HIF-1α downregulation led to hyperopia, whereas upregulation resulted in myopia. In human subjects, near work, a risk factor for myopia, significantly decreased choroidal blood perfusion, which might cause scleral hypoxia.
Interpretation: Our study implicated the HIF-1α signaling pathway in promoting human myopia through mediating interactions between genetic and environmental factors.
Funding: National Natural Science Foundation of China grants; Natural Science Foundation of Zhejiang Province.
Keywords: Genetic and environmental interactions; HIF-1α; Myopia; Myopia risk genes; Near work; Sclera.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Figures




References
-
- Dolgin E. The myopia boom. Nature. 2015;519:276–278. - PubMed
-
- Morgan IG, Ohno-Matsui K, Saw SM. Myopia. Lancet. 2012;379:1739–1748. - PubMed
-
- Grossniklaus HE, Green WR. Pathologic findings in pathologic myopia. Retina. 1992;12:127–133. - PubMed
-
- Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45:2873–2878. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

