A fork in the road: Where homologous recombination and stalled replication fork protection part ways
- PMID: 32653304
- PMCID: PMC8082280
- DOI: 10.1016/j.semcdb.2020.07.004
A fork in the road: Where homologous recombination and stalled replication fork protection part ways
Abstract
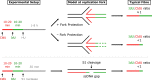
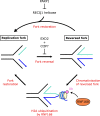
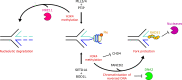
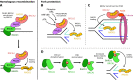
In response to replication hindrances, DNA replication forks frequently stall and are remodelled into a four-way junction. In such a structure the annealed nascent strand is thought to resemble a DNA double-strand break and remodelled forks are vulnerable to nuclease attack by MRE11 and DNA2. Proteins that promote the recruitment, loading and stabilisation of RAD51 onto single-stranded DNA for homology search and strand exchange in homologous recombination (HR) repair and inter-strand cross-link repair also act to set up RAD51-mediated protection of nascent DNA at stalled replication forks. However, despite the similarities of these pathways, several lines of evidence indicate that fork protection is not simply analogous to the RAD51 loading step of HR. Protection of stalled forks not only requires separate functions of a number of recombination proteins, but also utilises nucleases important for the resection steps of HR in alternative ways. Here we discuss how fork protection arises and how its differences with HR give insights into the differing contexts of these two pathways.
Keywords: BRCA1; BRCA2; Fork reversal; Homologous recombination; RAD51; Replication fork protection.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
The authors declare they have no financial or personal relationships with other people or organizations that could inappropriately influence this work.
Figures



References
-
- Neelsen K.J., Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015;16(4):207–220. - PubMed
-
- Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K., Herrador R., Jacobs H., Cortez D., Ciccia A., Penengo L., Lopes M. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 2017;67(5):882–890. e5. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

