Detailed Phenotyping and Therapeutic Strategies for Intronic ABCA4 Variants in Stargardt Disease
- PMID: 32653833
- PMCID: PMC7352060
- DOI: 10.1016/j.omtn.2020.06.007
Detailed Phenotyping and Therapeutic Strategies for Intronic ABCA4 Variants in Stargardt Disease
Abstract
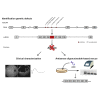
Stargardt disease is a progressive retinal disorder caused by bi-allelic mutations in the ABCA4 gene that encodes the ATP-binding cassette, subfamily A, member 4 transporter protein. Over the past few years, we and others have identified several pathogenic variants that reside within the introns of ABCA4, including a recurrent variant in intron 36 (c.5196+1137G>A) of which the pathogenicity so far remained controversial. Detailed clinical characterization of this variant confirmed its pathogenic nature, and classified it as an allele of intermediate severity. Moreover, we discovered several additional ABCA4 variants clustering in intron 36. Several of these variants resulted in aberrant splicing of ABCA4, i.e., the inclusion of pseudoexons, while the splicing defects caused by the recurrent c.5196+1137G>A variant strongly increased upon differentiation of patient-derived induced pluripotent stem cells into retina-like cells. Finally, all splicing defects could be rescued by the administration of antisense oligonucleotides that were designed to specifically block the pseudoexon insertion, including rescue in 3D retinal organoids harboring the c.5196+1137G>A variant. Our data illustrate the importance of intronic variants in ABCA4 and expand the therapeutic possibilities for overcoming splicing defects in Stargardt disease.
Keywords: ABCA4; Stargardt disease; antisense oligonucleotides; iPSC; intronic mutations; organoids; photoreceptors; retina; splicing; stem cells.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




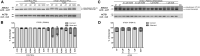
References
-
- Allikmets R., Singh N., Sun H., Shroyer N.F., Hutchinson A., Chidambaram A., Gerrard B., Baird L., Stauffer D., Peiffer A. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 1997;15:236–246. - PubMed
-
- Cornelis S.S., Bax N.M., Zernant J., Allikmets R., Fritsche L.G., den Dunnen J.T., Ajmal M., Hoyng C.B., Cremers F.P. In Silico Functional Meta-Analysis of 5,962 ABCA4 Variants in 3,928 Retinal Dystrophy Cases. Hum. Mutat. 2017;38:400–408. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

