ZFHX3 mutation as a protective biomarker for immune checkpoint blockade in non-small cell lung cancer
- PMID: 32653938
- PMCID: PMC10992006
- DOI: 10.1007/s00262-020-02668-8
ZFHX3 mutation as a protective biomarker for immune checkpoint blockade in non-small cell lung cancer
Abstract
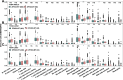
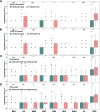
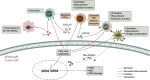
To date, immunotherapy has opened a new chapter in the treatment of lung cancer. Precise biomarkers can help to screen subpopulations of lung cancer to provide the best treatment. Multiple studies suggest that specific gene mutations may be predictive markers in guiding non-small cell lung cancer (NSCLC) immune checkpoint inhibitor (ICI) treatment. A published immunotherapy cohort with mutational and survival data for 350 NSCLC patients was used. First, the mutational data of the immunotherapy cohort were used to identify gene mutations related to the prognosis of ICI therapy. The immunotherapy cohort and TCGA-NSCLC cohort were further studied to elucidate the relationships between specific gene mutations and tumor immunogenicity, antitumor immune response capabilities, and immune cell and mutation counts in the DNA damage response (DDR) pathway. In the immunotherapy cohort (N = 350), ZFHX3 mutations were an independent predictive biomarker for NSCLC patients receiving ICI treatment. Significant differences were observed between ZFHX3-mutant (ZFHX3-MT) and ZFHX3-wild type (ZFHX3-WT) patients regarding the overall survival (OS) time (P < 0.001, HR = 0.26, 95% Cl 0.17-0.41). ZFHX3-MT is significantly associated with higher tumor mutation burden (TMB) and neoantigen load (NAL), and ZFHX3-MT positively correlates with known immunotherapy response biomarkers, including T-cell infiltration, immune-related gene expression, and mutation counts in the DDR pathway in NSCLC. ZFHX3-MT is closely related to longer OS in NSCLC patients treated with ICIs, suggesting that ZFHX3 mutations be used as a novel predictive marker in guiding NSCLC ICI treatment.
Keywords: Biomarker; Immune checkpoint inhibitor; Non-small cell lung cancer; ZFHX3.
Conflict of interest statement
The authors declare that they have no conflict interests.
Figures




References
-
- Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015. JAMA Oncol. 2017;3:524. doi: 10.1001/jamaoncol.2016.5688. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

