Peste des petits ruminants in Africa: a review of currently available molecular epidemiological data, 2020
- PMID: 32653984
- PMCID: PMC7497342
- DOI: 10.1007/s00705-020-04732-1
Peste des petits ruminants in Africa: a review of currently available molecular epidemiological data, 2020
Abstract
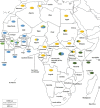
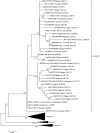
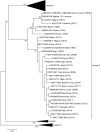
Small ruminants (e.g., sheep and goats) contribute considerably to the cash income and nutrition of small farmers in most countries in Africa and Asia. Their husbandry is threatened by the highly infectious transboundary viral disease peste des petits ruminants (PPR) caused by peste-des-petits-ruminants virus (PPRV). Given its social and economic impact, PPR is presently being targeted by international organizations for global eradication by 2030. Since its first description in Côte d'Ivoire in 1942, and particularly over the last 10 years, a large amount of molecular epidemiological data on the virus have been generated in Africa. This review aims to consolidate these data in order to have a clearer picture of the current PPR situation in Africa, which will, in turn, assist authorities in global eradication attempts.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Amarasinghe GK, Bào Y, Basler CF, Bavari S, Beer M, Bejerman N, Blasdell KR, Bochnowski A, Briese T, Bukreyev A, Calisher CH, Chandran K, Collins PL, Dietzgen RG, Dolnik O, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Fang Q, Formenty P, Fouchier RAM, Ghedin E, Harding RM, Hewson R, Higgins CM, Hong J, Horie M, James AP, Jiāng D, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lee B, Leroy EM, Li M, Maisner A, Mühlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Pearson MN, Randall RE, Revill PA, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Smither SJ, Song Q, Stone DM, Takada A, Terregino C, Tesh RB, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Volchkov VE, Wahl-Jensen V, Walker PJ, Wang B, Wang D, Wang F, Wang LF, Werren JH, Whitfield AE, Yan Z, Ye G, Kuhn JH. Taxonomy of the order Mononegavirales: update. Arch Virol. 2017;162:2493–2504. - PMC - PubMed
-
- Bailey D, Banyard A, Dash P, Ozkul A, Barrett T. Full genome sequence of peste des petits ruminants virus, a member of the Morbillivirus genus. Virus Res. 2005;110:119–124. - PubMed
-
- Diallo A, Barrett T, Barbron M, Meyer G, Lefèvre PC. Cloning of the nucleocapsid protein gene of peste-des-petits-ruminants virus: relationship to other morbilliviruses. J Gen Virol. 1994;75:233–237. - PubMed
-
- Meyer G, Diallo A. The nucleotide sequence of the fusion protein gene of the peste des petits ruminants virus: the long untranslated region in the 5'-end of the F-protein gene of morbilliviruses seems to be specific to each virus. Virus Res. 1995;37:23–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

